Supplemental Data. Mettler et al. Plant Cell (2014) 10 ...€¦ · 5/8/2014 · Supplemental Data....
Transcript of Supplemental Data. Mettler et al. Plant Cell (2014) 10 ...€¦ · 5/8/2014 · Supplemental Data....

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Harvest tube
Oxygen electrode
Acid feedBase feedMedium feedGas out
Stirrer
Turbidimeter
Pressure sensor
Temperature sensor
Closed metal tube
pH probe
Gas in
Cooling water out
Cooling water in
Light intensity probe
Closed glass tube
Light shell
SpargerSampling port
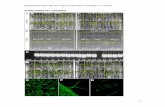
Supplemental Figure 1. Schematic Showing the Components of the Bioreactors.
5-litre bioreactors BIOSTAT®B-DCU (Sartorius Stedim, Germany) were used to grow continuous cultures. The reactorswere bubbled with 200 cm3*min-1 of altered 5% CO2 in air and are stirred at 50 rpm with three impellers. Cell density wasmonitored by a turbidimeter, and when set threshold was exceeded (equivalent to 3-4*106 cells*ml-1), autoclave-sterilised medium was pumped automatically into the bioreactor to dilute the culture. The entire bioreactor was set onscales, and when an increase in weight was detected from the addition of fresh medium, culture was pumped out via theharvest tube to a sterile 20 L carboy. The pH of the culture was constantly monitored, and if the pH dropped below 6.95,sterile 1 M KOH was slowly pumped into the bioreactor until the pH was within 0.05 units of 7.00. If the pH increasesabove 7.05, sterile 1 M HCl was pumped into the bioreactor. Temperature was also controlled, and regulated by watercirculating within an outer mantle, in addition to water used to cool the light system. If the pressure within the headspaceexceeds a set value, an alarm was triggered. All of these parameters, and others, like the dissolved oxygen concentration,are logged in the MFCS/win software, which also controls all parameters of the bioreactor, including light in conjunctionwith a second software program, Quattro Color Light is supplied from two LED half-shells and is focused with individuallenses to the centre of the bioreactor, to minimise light gradients within the bioreactor.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
0
10
20
30
40
50
60
70
80
90
0 500 1000 1500 2000
Re
lati
ve E
TR
Light intensity [µmol*m-2*s-1]
Treatment -60 min
Treatment 0 min
Treatment 60 min
Treatment 120 min
Treatment 240 min
Treatment 480 min
0
30
60
0 50 100 150 200
0
10
20
30
40
50
60
70
80
90
0 500 1000 1500 2000
Re
lati
ve E
TR
Light intensity [µmol*m-2*s-1]
Control -60 min
Control 0 min
Control 60 min
Control 120 min
Control 240 min
Control 480 min
0
30
60
0 50 100 150 200
A
B
Supplemental Figure 2. Light Saturation Curves of C. reinhardtii CC-1690 Cells Under Low Light (A) and during the LightShift Experiment (B).
(A) In the control conditions, light was kept at a light intensity of 41 µmol photons*m-2*s-1 for the course of theexperiment. Relative ETR was measured at -60 min, 0 min, 60 min, 120 min, 240 min and 480 min during the time course(n = 3 ± SD).(B) In the treatment conditions, light intensity was shifted from 41 to 145 µmol photons*m-2*s-1 at time point zero.Relative ETR was measured one hour before the light shift (-60min), just before the light shift (0 min) and 60 min, 120min, 240 min and 480 min after the light shift (n = 3 ± SD).Arrows in the inserts indicate growth light intensities.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
-120 -60 0 60 120 180 240 300 360 420 480 540
Time after light shift [min]
Transcripts
Proteins
Metabolites(LC-MS/MS)
Metabolites(GC-MS, LC-MS)
-120 -60 0 60 120 180 240 300 360 420 480 540
Time after light shift [min]
145 µmol photons*m-2*s-1
41µmol photons*m-2*s-1
A
B
Polysomeloading
Transcripts
Proteins
PS parameters
Metabolites(LC-MS/MS)
Metabolites(GC-MS, LC-MS)
Polysomeloading
Supplemental Figure 3. Experimental Setup.
C. reinhardtii CC-1690 cells were grown continuously at 24°C, 5% CO2 and 41 µmol photons*m-2*s-1 for three days beforethe start of the experiment to assure steady-state conditions. The functional levels analysed are given on the left and theunits of each level on the right side of the graph (PS, photosynthesis; OD, optical density). The time points the sampleswere taken for the different functional levels are indicated by vertical lines. Two (proteins and metabolites, additionallythree and two technical replicates were analysed, respectively) or three (other functional levels) bioreactor runs wereperformed. Metabolites of the central metabolism were measured absolutely and relatively, respectively, with LC-MS/MSand GC-MS and lipids were measured relatively with LC-MS (for details, see Methods).(A) In the control conditions, light was kept at the initial light intensity for the course of the experiment.(B) In the treatment conditions, light intensity was shifted from 41 (light yellow) to 145 µmol photons*m-2*s-1 (darkyellow) at time point zero.
Relative to total RNA
Relative to internal standard
Relative to OD
Concentration per cell volume
Relative to OD
Relative to total ribosomes
Relative to total RNA
Relative to internal standard
Relative to OD
Concentration per cell volume
Relative to OD
Relative to total ribosomes
PS parameters

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Time after light shift [min]
*** *** *** ******
*** *** ***
Supplemental Figure 4. Oxygen Concentration in the Outlet Air of the Bioreactor.
Oxygen concentration in the outlet air of the open-system bioreactor after the increase in light intensity at time pointzero (dashed line). Additionally, the medium was pumped 2.4 times faster at the higher light intensity (n = 24 ± SD,Student´s t-test: three asterisks, p<0.001).
20.0
21.0
22.0
23.0
24.0
-60 min 60 min 120 min 180 min 240 min 300 min 360 min 420 min 480 min
low light higher light
Oxy
gen
evo
luti
on
[%
]

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
0
20
40
60
80
100
-120 -60 0 60 120 180 240 300 360 420 480 540
Tota
l pro
tein
[µ
g*m
l-1]
Time after light shift [min]
Supplemental Figure 5. Total Protein Content.
C. reinhardtii CC1690 cells were grown in a bioreactor at 24°C, 5% CO2 and 41 µmol photons*m-2*s-1. At time point zero the light was either kept at theinitial light intensity (open cycles, n = 2 ± SD) or shifted to 145 µmol photons*m-2*s-1 (filled cycles, n = 2 ± SD) at time point zero. Total protein contentwas measured by Bradford assay as described in Methods.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 6. Venn Diagrams Showing Numbers of Significantly Changed Transcripts (A, C) and Proteins (B, D).
Numbers in the Venn diagrams indicate significantly changed transcripts and proteins determined by using an ANOVArefined by a contrast analysis with p-value < 0.05. Additionally, a relevance threshold (-1 ≥ log2 (fold change) ≥ 1) wasapplied to the transcript data set. C, control; T, treatment.(A and B) Venn diagrams showing the numbers of transcripts (A) and proteins (B) significantly changed after the light shiftat early (red) and late time points (green) compared to the control (blue). The control is the combination of the timepoints before the light shift and the 480 min time point of the untreated control bioreactor. Transcripts and proteins thatdiffer within the control are used to check for changes over time not related to the light shift (blue, for more details, see Cand D).(C and D) Venn diagrams showing the numbers of transcripts (C) and proteins (D) significantly changing due to an effectusing separate bioreactors (blue) and a time effect (green), respectively. Transcripts and proteins that significantly differbetween the control and treatment bioreactors in time points before the light shift can be explained by the bioreactoreffect. Changes over time in the untreated control bioreactor can be explained by the time effect. Both, the time andbioreactor effect are independent of the effect by the shift in light.
C D
A B

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Con
trol_0m
inup
Con
trol_48
0minup
0minup
40m
inup
120m
inup
480m
inup
Con
trol_0m
indo
wn
Con
trol_48
0mindo
wn
0mindo
wn
40m
indo
wn
120m
indo
wn
480m
indo
wn
PSmajor CHO metabolismminor CHO metabolismglycolysisfermentationgluconeogenese/ glyoxylate cycleOPPTCA / org transformationmitochondrial electron transport / ATP synthesiscell wall
lipid metabolismN-metabolismamino acid metabolismS-assimilationmetal handlingsecondary metabolismhormone metabolismCo-factor and vitamine metabolismtetrapyrrole synthesisstressredoxpolyamine metabolismnucleotide metabolismBiodegradation of XenobioticsC1-metabolismmiscRNA
DNAprotein
signallingcell
micro RNA, natural antisense etcdevelopmenttransport
not assigned
Mineral Nutrition
PS
cell wall
cell wall.cell wall proteins
secondary metabolism
RNARNA.processing
proteinprotein.aa activationprotein.synthesisprotein.targeting
protein.targeting.mitochondria
protein.targeting.secretory pathway
protein.degradation
protein.degradation.serine protease
protein.degradation.ubiquitin
protein.degradation.ubiquitin.proteasom
protein.folding
protein.folding.chaperones and co-chaperones
cell
cell.motilitycell.motility.eukaryotes
cell.motility.eukaryotes.intraflagellar transport
cell.motility.eukaryotes.intraflagellar transport.IFT particle protein
cell.motility.eukaryotes.intraflagellar transport.IFT particle protein.complex Bcell.motility.eukaryotes.axonemal dyneins
cell.motility.eukaryotes.axonemal dyneins.inner armcell.motility.eukaryotes.axonemal dyneins.inner arm.monomeric species
cell.motility.eukaryotes.flagellar associated proteins
transport
transport.ABC transporters and multidrug resistance systems
not assigned
not assigned.unknown
0
-3
3
-2
-1
1
2
Con
trol_
0min
up
Con
trol_
480m
inup
0min
up 40m
inup 12
0min
up48
0min
upC
ontro
l_0m
indo
wn
Con
trol_
480m
indo
wn
0min
dow
n40
min
dow
n
120m
indo
wn
480m
indo
wn
PS
ma
jor
CH
O m
eta
bo
lism
min
or
CH
O m
eta
bolis
mgly
coly
sis
ferm
en
tation
glu
coneogenese/ gly
oxyla
te c
ycle
OP
PT
CA
/ o
rg tra
nsfo
rma
tio
nm
ito
cho
nd
ria
l e
lectr
on
tra
nsp
ort
/ A
TP
syn
the
sis
ce
ll w
all
lipid
meta
bolis
mN
-meta
bolis
ma
min
o a
cid
me
tabo
lism
S-a
ssim
ilatio
nm
eta
l h
and
ling
se
co
nda
ry m
eta
bo
lism
horm
one m
eta
bolis
mC
o-f
acto
r a
nd
vita
min
e m
eta
bo
lism
tetr
apyrr
ole
syn
the
sis
str
ess
redo
xpoly
am
ine m
eta
bolis
mnucle
otide m
eta
bolis
mB
iodegra
dation o
f X
enobio
tics
C1-m
eta
bolis
mm
isc
RN
A
DN
Apro
tein
sig
nalli
ng
ce
ll
mic
ro R
NA
, n
atu
ral a
ntise
nse e
tcdevelo
pm
ent
tra
nsp
ort
not assig
ned
Min
era
l N
utr
itio
n
PS
cell
wa
ll
ce
ll w
all.
ce
ll w
all
pro
tein
s
se
co
nda
ry m
eta
bo
lism
RN
AR
NA
.pro
ce
ssin
g
pro
tein
pro
tein
.aa
activa
tio
np
rote
in.s
yn
the
sis
pro
tein
.ta
rgeting
pro
tein
.ta
rge
tin
g.m
ito
cho
nd
ria
pro
tein
.ta
rge
tin
g.s
ecre
tory
path
wa
y
pro
tein
.de
gra
da
tio
n
pro
tein
.de
gra
da
tio
n.s
erin
e p
rote
ase
pro
tein
.de
gra
da
tio
n.u
biq
uitin
pro
tein
.de
gra
da
tio
n.u
biq
uitin
.pro
tea
so
m
pro
tein
.fo
ldin
g
pro
tein
.fo
ldin
g.c
hap
ero
ne
s a
nd
co
-ch
ap
ero
ne
s
cell
ce
ll.m
otilit
yce
ll.m
otilit
y.e
uka
ryo
tes
cell.
mo
tilit
y.e
ukary
ote
s.intr
aflag
ella
r tr
anspo
rt
ce
ll.m
otilit
y.e
uka
ryo
tes.in
tra
fla
ge
llar
tra
nsp
ort
.IF
T p
art
icle
pro
tein
ce
ll.m
otilit
y.e
uka
ryo
tes.in
tra
fla
ge
llar
tra
nsp
ort
.IF
T p
art
icle
pro
tein
.co
mple
x B
cell.
mo
tilit
y.e
ukary
ote
s.a
xo
ne
ma
l d
yn
ein
s
ce
ll.m
otilit
y.e
uka
ryo
tes.a
xon
em
al d
yn
ein
s.in
ne
r a
rmce
ll.m
otilit
y.e
uka
ryo
tes.a
xon
em
al d
yn
ein
s.in
ne
r a
rm.m
ono
meric s
pe
cie
s
cell.
mo
tilit
y.e
ukary
ote
s.fla
gella
r a
ssocia
ted p
rote
ins
tra
nsp
ort
tra
nspo
rt.A
BC
tra
nspo
rte
rs a
nd
mu
ltid
rug r
esis
tance s
yste
ms
not assig
ned
not assig
ned.u
nknow
n
0-3 3-2 -1 1 2
Supplemental Figure 7. Comparison of Significant Over-represented Functional Gene Categories (and Sub-Categories) of Different Time Points during the Light Shift Applied to C. reinhardtii CC-1690 Cells Determined by PageMan.
The values are representative of the log2 transformed values of three individual hybridizations and classed according to the MapMan classifications (Thimmet al., 2004; Usadel et al., 2005). Over-representation of classifications was assessed via Fisher test (p-value < 0.05; Usadelet al., 2006). Red indicates an increase whereas blue indicates a decrease (see color scale). Controls where light was kept at the initial light intensities are shaded in grey. Note, first, the overrepresentation for photosynthesis genes at 120 min after the light shift, second, the overrepresentation of genes involved in protein targeting and degradation and third, the overrepresentation of decreased genes involved in cell mobility.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
A
B
C
Transcripts
Proteins
Metabolites
-10
-5
0
5
10
-120 -60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
PSBS1
BAG6
NRX3
LHCSR1
Cre08.g384650
CCP2
_ADH5
Cre02.g106450
LCI1
Cre16.g681400
-1.5
-1
-0.5
0
0.5
1
1.5
-120 -60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
PDX1
VIPP1
_CPK3
Cre07.g344400
METE
SHMT2
ATP5
PSBR
PSBW
_ICL2
D
Lipids
-3
-2
-1
0
1
2
3
-120 -60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
RuBP
GAP
Alanine
S7P
Threonic acid
F6P
Serine
Threonine
ADPG
DHAP
-3
-2
-1
0
1
2
3
-120 -60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
DGDG 34:5
DGDG 34:6
DGTS 32:0
DGTS 34:8
DGTS 36:2
DGTS 36:7
DGTS 36:8
MGDG 34:1
MGDG 34:3
TAG 54:9
Supplemental Figure 8. Top-10 Log2-fold Changes of Transcripts (A), Proteins (B) Metabolites (C) and Lipids (D) during the Light Shift Applied to C. reinhardtii CC-1690 Cells.
Log2-fold changes were calculates for each measured time point after the light was increased from 41 to 145 µmol photons*m-
2*s-1 relative to the average of the -60 min and 0 min time points before the light was increased. For further details of analyses see Methods. (A, C and D) The transcripts (A), metabolites (C) and lipids (D) with the ten most extreme log2-fold changes at any time point at the higher light intensity are shown.(B) Proteins with the ten most extreme average log2-fold changes at the higher light intensity are shown.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
-1
-0.8
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
RPS29
RPS10
RPS14
RPS15
RPS17
RPS18
RPS19
RPS21
RPS4
RPS5
RPS3a
RPS6
RPS7
-1
-0.8
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
PRPS13
PRPS15
rps2
PRPS20
rps3
PRPS5
PSRP-1
rps7
rps9
PSRP3
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
rbcL
RPI1
PRK1
RCA1
RBCS1
FBP1
TRK1
SBP1
PGK1
GAP3
TPIC
FBA1+FBA3-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
LHCB4
LHCBM3
LHCBM2
LHCBM7
LHCB5
LHCBM4
LHCBM6
LHCBM1
LHCBM5
LHCBM8
-1
-0.8
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
PSBQ
PSBP1
PSBO
psbD
psbB
psbE
CPLD45
PSBR
psbA
psbC
PSBP6
PSBW
APR
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
LHCA9
LHCA3
LHCA1
LHCA4
LHCA6
LHCA7
LHCA5
LHCA2
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
PSAG
PSAH
Cre10.g433950
PSAE
PSAL
psaA
PSAD
psaB
psaC
PSAK
PSAN
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
petD
petA
PETC
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
ATPC
atpA
ATPG
atpB
ATPD
-1
-0.8
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
PRPL1
PRPL7
rpl14
PRPL18
PRPL19
rpl2
PRPL24
Cre01.g050308
PRPL31
PRPL4
rpl5
PRPL6
-1
-0.8
-0.6
-0.4
-0.2
0
0.2
0.4
0.6
-60 0 60 120 180 240 300 360 420 480 540
Log 2
-fo
ld c
han
ge
Time after light shift [min]
RPL10
RPL11
RPL13
RPL22
RPL26
RPL27
RPL28
RPL3
RPL32
RPL35
RPL36
RPL37
RPL5
RPL23a
RPL27a
RPL6
RPL7
RPP2
LHCII subunits (bin 1.1.1.1)
PSII subunits (bin 1.1.1.2)
LHCI subunits (bin 1.1.2.1)
PSI subunits (bin 1.1.2.2)
Cytochrome b6f complex subunits (bin 1.1.3)
ATPase subunits (bin 1.1.4)
Calvin-Benson cycle enzymes (bin 1.3)
Ribosome small subunits (prokaryotic) (bin 29.2.1.1.1.1)
Ribosome large subunits (prokaryotic) (bin 29.2.1.1.1.2)
Ribosome small subunits (eukaryotic) (bin 29.2.1.2.1)
Ribosome large subunits (eukaryotic) (bin 29.2.1.2.2)
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
Supplemental Figure 9. Protein Levels in C. reinhardtii CC-1690Cells During the Light Shift Experiment within the FunctionalCategories “Light Reaction”, “CBC” and “Ribosomes”.
Kinetics of individual proteins of selected functional categoriesbased on MapMan ontology (bin number indicated inparentheses). Central tendencies are shown in Figure 4. Asterisksmark significantly changed proteins groups (p-value<0.05). Toallow good visualization of all CBC proteins, data for FBA1 andFBA3 were combined as FBA1 is a minor component of aldolaseactivity (Blaby et al. 2013; see text and Supplemental Table 1).Asterisks indicate proteins that change significantly (ANOVA p-value<0.05, after p-value correction for multiple sampling byBenjamini-Hochberg). For further details of analyses see Methods.
+RBCS2

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 10. 77K Fluorescence Emission Spectra during Light Shift.
For comparability of the different emission signals, the maximum fluorescence emission of PSII at 687 nm wavelength wasnormalized to one.(A) In the control conditions , C. reinhardtii CC-1690 cells were exposed to a light intensity of 41 µmol photons*m-2*s-1 forthe course of the experiment. A 77K fluorescence emission spectrum was measured at -60 min, 0 min, 60 min, 120 min,240 min and 480 min during the time course.(B) In the treatment conditions, light intensity was shifted from 41 to 145 µmol photons*m-2*s-1 at time point zero. A 77Kfluorescence emission spectrum was measured one hour before the light shift (-60min), just before the light shift (0 min)and 60 min, 120 min, 240 min and 480 min after the light shift.
0
0.5
1
1.5
660 680 700 720 740 760 780 800
Ch
loro
ph
yll-
a f
luo
resc
en
ce e
mis
sio
n
Wave length [nm]
Control -60 min
Control 0 min
Control 60 min
Control 120 min
Control 240 min
Control 480 min
0
0.5
1
1.5
660 680 700 720 740 760 780 800
Ch
loro
ph
yll-
a f
luo
resc
en
ce e
mis
sio
n
Wave length [nm]
Treatment -60 min
Treatment 0 min
Treatment 60 min
Treatment 120 min
Treatment 240 min
Treatment 480 min
A
B

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
0
0.2
0.4
0.6
0.8
1
-60 0 5 10 20 40 60 120 240 480
AEC
[(A
TP+0
.5*A
DP
)/(A
TP+A
DP
+AM
P)]
Time after light shift [min]
Supplemental Figure 11. Adenylate Energy Charge (AEC) during Light Shift.
From ATP measured spectrophotometrically and ADP and AMP measured via LC-MS/MS in C. reinhardtii CC-1690 cells,the AEC was calculated for each measured time point in the control samples (white bars), where light was kept at 41 µmolphotons*m-2*s-1, and the treatment samples (black bars), where light was shifted to 145 µmol photons*m-2*s-1 at timepoint zero (n = 4 ± SD).

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 12. CBC Intermediate Levels Change with Increasing Light Intensity.
C. reinhardtii CC-1690 cells were grown in a bioreactor at 24°C, 5% CO2 and 46 µmol photons*m-2*s-1. Before harvestingand measurement cells were exposed for one hour to darkness or to the indicated light intensities.(A) CBC intermediate levels in the dark and five different light intensities were measured by LC-MS/MS (n = 8, ± SD).(B) Metabolite ratios for selected CBC intermediates based on metabolite levels shown in A.(C) Net photosynthesis rates in Chlamydomonas cells were measured after the addition of 10 mM sodium bicarbonate atthe corresponding light intensities in an external closed cuvette using an optical oxygen sensor (n = 4, ± SD).(D) Gibbs free energy of reaction (rG) of Calvin-Benson cycle enzymes. For calculations, standard free energies (rG
0´)from Bassham and Krause (1969) and the formula rG =rG
0´+R*T*ln(MAR) were used, where the mass action ratio(MAR) is the reaction quotient that was calculated based on the CBC intermediate levels shown in A. The followingassumption were made: a [CO2] under 5% CO2 conditions was assumed to be 0.202M. b ATP/ADP=3 (light), ATP/ADP=0.5(dark) (Gardestrom and Wigge, 1988), c NADPH/NADP+=1 (light), NADPH/NADP+=0.5 (dark) (Heineke et al., 1991), d
[Pi]=0.002M (Pratt et al., 2009). e The first TRK reaction was assumed to be at equilibrium to estimate the in vivoconcentration of E4P. The fact that rG of SBA is also close to zero in the light supports the assumption that the first TRKand SBA catalysed reactions are close to equilibrium and either reaction could have been used to estimate E4Pconcentration, with approximately the same result. This was not the case for cells exposed to dark. The TRK and SBAreaction are likely not at equilibrium in the dark and E4P estimation is difficult. f The RPE reaction was assumed to be atequilibrium to estimate individual levels of X5P and Ru5P, which are stereoisomers and not distinguishable with LS-MS/MS. The rG of RPI being close to zero supports this assumption.
3PGA
0 20 40 60 80 100 120 140 160
Co
nce
ntr
ati
on
[µ
M]
0
200
400
600
800
1000
DHAP
0 20 40 60 80 100 120 140 160
Co
nce
ntr
ati
on
[µ
M]
0
100
200
300
400
500
600
700
F6P
0 20 40 60 80 100 120 140 160
Co
nce
ntr
ati
on
[µ
M]
0
200
400
600
800
1000
SBP
0 20 40 60 80 100 120 140 160
Co
nce
ntr
ati
on
[µ
M]
0
50
100
150
200
250
Xu5P+Ru5P
Light intensity [µmol photons*m-2*s-1]
0 20 40 60 80 100 120 140 160
Co
nce
ntr
ati
on
[µ
M]
0
10
20
30
40
50
60
FBP
0 20 40 60 80 100 120 140 160
0
50
100
150
200
GAP
0 20 40 60 80 100 120 140 160
0
10
20
30
40
50
R5P
0 20 40 60 80 100 120 140 160
0
10
20
30
40
50
RuBP
Light intensity [µmol photons*m -2*s-1]
0 20 40 60 80 100 120 140 160
0
500
1000
1500
2000
S7P
0 20 40 60 80 100 120 140 160
0
200
400
600
800
1000
1200
1400
1600
1800
-20
0
20
40
60
80
100
120
140
0 20 40 60 80 100 120 140 160
Ne
t p
ho
tosy
nth
esis
rat
e[µ
mo
l O2*h
-1*m
g ch
loro
ph
yll-1
]
Light intensity [µmol photons*m-2*s-1]
-60
-50
-40
-30
-20
-10
0
10
20Δ
Gib
bs
free
en
ergy
of
reac
tio
n [
kJ/m
ol]
dark 20µE 34µE 46µE 94µE 143µE
A B
C
D
n.d.
TPI FBA FBPased TRKe SBA SBPased TRK RPEf RPI PRKbPGK+GAPDH
b,c,d
Rubis-coa
0
0.02
0.04
0.06
0.08
0
0.2
0.4
0.6
0.8
1
1.2
0 20 40 60 80 100 120 140 160
Light intensity[µmol photons*s-1*m-2]
DHAP/3PGA(left y-axis)GAP/3PGA(right y-axis)
0
5
10
15
20
25
30
0 20 40 60 80 100 120 140 160
Light intensity[µmol photons*s-1*m-2]
F6P/FBP
S7P/SBP

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 13. Heatmap of Lipid Levels during Light Shift in C. reinhardtii CC-1690 Cells.
Log2-normalized data are shown where for each time point the value was centred to the median of the correspondinglipid. Color key indicates lipid accumulation value, blue: lowest, red: highest. Arrows indicate lipids that changesignificantly (ANOVA p-value<0.05, after p-value correction for multiple sampling by Benjamini-Hochberg).

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 14. Amino Acids and Organic Acids.
C. reinhardtii CC1690 cells were grown in a bioreactor at 24°C, 5% CO2 and 41 µmol photons*m-2*s-1
(open circles) or shifted to 145 µmol photons*m-2*s-1 at time point zero (filled circles). Metabolites were separated and detected by LC-MS/MS (isocitrate, malate, aconitate, 2OG) (n = 4 ±SD). Metabolite levels are given as concentrations (µM; see Methods). Citrate, succinate, fumarate, glutamate, alanine, phenylalanine, aspartate, serine and threonine were separated and detected by GC-MS (n = 3 ± SD) and given in fold-changes normalized to all reference time points (see Methods). Pairwise t-test between control and treatment was done and p-value correction for multiple sampling by Bonferroni correction (one asterisk, p < 0.05; two asterisks, p < 0.01; three asterisks, p < 0.001). ANOVA analysis was done and p-value correction for multiple sampling by Benjamini-Hochberg correction (dagger, p > 0.05; double dagger, p < 0.05). Graphs are based on data given in Supplemental Dataset 1.
Time after light shift [min]
***
Time after light shift [min]
***
**
***
‡
‡
†
†
†
†
††
†
‡
†
†
†
†
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Citrate
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Glutamate
††
0
20
40
60
80
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Co
nce
ntr
atio
n [
µM
] Isocitrate
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Alanine
†
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Succinate
0
2
4-1
20
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Phenylalanine
****** ***
‡
††
0
2000
4000
6000
8000
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Co
nce
ntr
atio
n [
µM
] Malate
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Asparate
0
4
8
12
16
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Co
nce
ntr
atio
n[µ
M] Aconitate
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
cah
nge
Serine
††
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Fumarate
0
2
4
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Fold
chan
ge
Threonine
0
100
200
300
400
-12
0
-60 0
60
12
0
18
0
24
0
30
0
36
0
42
0
48
0
54
0
Co
nce
ntr
atio
n [
µM
] 2OG
†
†
†
‡

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
y = 0.2902x - 0.0964R² = 0.791
-2
-1.5
-1
-0.5
0
0.5
1
1.5
2
-4 -2 0 2 4
Log 2
-fo
ld c
han
ge o
f p
rote
in (
aver
age
ove
r ti
me)
Log2-fold change of transcripts (average over time)
y = -0.3997x + 0.0707R² = 0.3637
-2
-1.5
-1
-0.5
0
0.5
1
1.5
2
-4 -2 0 2 4
Log 2
-fo
ld c
han
ge o
f p
rote
in (
aver
age
ove
r ti
me)
Log2-fold change of transcripts (average over time)
-1
-0.5
0
0.5
1
-120 -60 0 60 120 180 240 300 360 420 480 540
Time after light shift [min]
AST3 METE
_SAS1 DPA1
LEU3 Cre07.g344400
ASL4
-4
-2
0
2
4
-120 -60 0 60 120 180 240 300 360 420 480 540
Time after light shift [min]
-4
-2
0
2
4
-120 -60 0 60 120 180 240 300 360 420 480 540
Time after light shift [min]
RCA1
FBA1
FBP1
SBP1
PGK1
GAP3
-1
-0.5
0
0.5
1
-120 -60 0 60 120 180 240 300 360 420 480 540
Time after light shift [min]
RCA1 FBA1
FBP1 SBP1
PGK1 GAP3
Log 2
-fo
ld c
han
ge c
om
par
ed c
on
tro
lsLo
g 2-f
old
ch
ange
co
mp
ared
co
ntr
ols
C D EAmino acid synthesis(bin 13.1)
Calvin-Benson cycle(bin 1.3)
Amino acid synthesis(bin 13.1)
Calvin-Benson cycle(bin 1.3)
Amino acid synthesis(bin 13.1)
Calvin-Benson cycle(bin 1.3)
Transcripts Proteins
Transcripts Proteins
y = 0.2163x - 0.0819R² = 0.2355
p-value<0.001
-2
-1.5
-1
-0.5
0
0.5
1
1.5
2
-4 -3 -2 -1 0 1 2 3 4
Log 2
-fo
ld c
han
ge o
f p
rote
in (
aver
age
ove
r ti
me)
Log2-fold change of transcripts (average over time)
all
t-/p+
t+/p+
t+/p-
t-/p-
t: transriptp: protein-: down+: up
Bin Bin name nPearson
correlationcoefficient
R2 p-value
11.1 lipid metabolism.FA synthesis and FA elongation 4 0.991 0.983 <0.001*
9.09 mitochondrial electron transport / ATP synthesis.F1-ATPase 5 0.947 0.896 0.004*
13.1 amino acid metabolism.synthesis 7 0.889 0.791 0.003
29.6.2 protein.folding 6 0.868 0.753 0.013*
1.1.1.2 PS.lightreaction.photosystem II.PSII polypeptide subunits 5 0.790 0.623 0.076
35.2 not assigned.unknown 10 0.756 0.572 0.008
29.2.1.1.1 protein.synthesis.ribosomal protein.prokaryotic.chloroplast 7 -0.604 0.365 0.134
1.3 PS.calvin cycle 6 -0.603 0.364 0.181
1.1.1.01 PS.lightreaction.photosystem II.LHC-II 6 -0.569 0.324 0.215
29.2.1 protein.synthesis.ribosomal protein 6 -0.499 0.249 0.293
1.1.2.2 PS.lightreaction.photosystem I.PSI polypeptide subunits 6 -0.394 0.155 0.424
31.6.1.10 cell.motility.eukaryotes.flagellar associated proteins 4 0.225 0.051 0.760
28.1.3 DNA.synthesis/chromatin structure.histone 5 -0.114 0.013 0.851
A B
Supplemental Figure 15. Correlation between Average Changes in Transcript and Protein Group Levels.
C. reinhardtii CC-1690 cells were grown in a bioreactor at 24°C, 5% CO2 and 41 µmol photons*m-2*s-1 and shifted to 145µmol photons*m-2*s-1 at time point zero. Levels of transcripts and protein groups were measured during the light shift asdescribed in Methods. Average log2-fold changes of samples exposed to higher light and changing significantly (ANOVA, p-value<0.05 after p-value correction by Benjamini-Hochberg method) are plotted against each other (A). Pearsoncorrelation coefficients between average changes of transcripts and protein groups were calculated for individualMapMan bins with a minimum of four members (B; red, positive correlation; blue, negative correlation; asterisk,significant correlations driven by single members). For two of these bins, details are shown in C-E.(C) The average of log2-fold change over time of transcripts is plotted versus the average log2-fold change of thedifferent protein groups and shown in grey as in A. In red, data for two MapMan bin is highlighted (bin 13.1 and 1.3).(D) The time course of the transcript levels belonging to the MapMan bins 13.1 and 1.3, respectively, are shown.(E) The time course of the protein groups belonging to the MapMan bins 13.1 and 1.3, respectively, are shown.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
atpA
atpB
rbcL
rps3rps9rpl14
rpl2rpl5 tufA
rpl20
y = 33.045e0.0101x
R² = 0.8278
5
50
500
5 50 500
Tran
scri
pt
abu
nd
ance
[p
g/µ
g to
tal R
NA
]
Protein abundance [µM]
Supplemental Figure 16. Correlation between Protein Levels and Transcript Levels for Plastid-encoded Genes in C.reinhardtii CC-1690 Cells.
Proteins and transcripts were quantified by the emPAI method (see Supplemental Table 1 and Methods) and microarrayanalysis with normalisation to synthetic spike-in RNAs (Kahlau and Bock, 2008), respectively. The ten plastid-encodedproteins which were quantified by emPAI showed a significant correlation (R² = 0.83, p = 0.0002) with their respectivetranscript levels (n = 3 ± SD).

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 17. Calculated Metabolite Ratios.
C. reinhardtii CC-1690 cells were grown in a bioreactor at 24°C, 5% CO2 and 41 µmol photons*m-2*s-1. At time point zerothe light was either kept at the initial light intensity (open circles) or shifted to 145 µmol photons*m-2*s-1 (filled circles) attime point zero. Based on the metabolite concentrations shown in Figure 6 and Supplemental Dataset 1, the ratios of3PGA/GAP (A), F6P/FBP (B) and S7P/SBP (C) were calculated from the average metabolite levels (n = 4).
A
B
C
0
200
400
600
800
1000
-120 -60 0 60 120 180 240 300 360 420 480 540
Rat
io 3
PG
A/G
AP
Time after light shift [min]
0
2
4
6
8
10
12
-120 -60 0 60 120 180 240 300 360 420 480 540
Rat
io F
6P
/FB
P
Time after light shift [min]
0
2
4
6
8
10
12
-120 -60 0 60 120 180 240 300 360 420 480 540
Rat
io S
7P
/SB
P
time after light shift [min]

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
1
10
100
1000
10000
100000
1 10 100 1000 10000 100000
[S]
at lo
w li
ght
[µM
]
Km [µM]
in vitro Km [µM]
in vivo Km low light [µM]
[S] = Km
Supplemental Figure 18. In Vitro and in Vivo Km-values versus Substrate Concentrations [S].
The substrate levels at low light of CBC reactions in C. reinhardtii CC-1690 cells (shown in Figure 6) are plotted against invitro Km (dark grey) (literature values, for details see Supplemental Table 6) and in vivo Km (light grey) calculated based onsubstrate saturation curves (shown in Figure 11). In Figure 12, an equal plot with substrate levels 20 min after the lightshift is shown. Arrows indicate >2-fold increases between in vitro and in vivo Km-values. Enzymes with their substrate inparentheses: a PGK (3PGA), b TRK (S7P), c Rubisco (RuBP), d FBA (DHAP), e SBA (DHAP), f oxidized SBPase (SBP), g reducedSBPase (SBP), h oxidized FBPase (FBP), i reduced FBPase (FBP), j RPI (R5P), k PRK (Ru5P), l SBA (E4P), m TPI (GAP), n FBA(GAP), o GAPDH (BPGA).
a
b bc c
d,e d ef g
h h i i
j
k k
l lmm
n n
o
In vitro Km [µM]In vivo Km in low light [µM][S]=Km

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
R² = 0.9724
0
100
200
300
400
500
0 0.2 0.4 0.6
Ban
d a
rea
[mm
2]
BSA protein amount [µg]
50kD
50kD
50kD
164 82 41 20.5 1 0.5 0.25 0.125
µl culture µg BSA protein
152 76 38 19 1 0.5 0.25 0.125
µl culture µg BSA protein
183 91 46 23 1 0.5 0.25 0.125
µl culture µg BSA protein
A B C
Supplemental Figure 19. Amount of RbcL Protein in C. reinhardtii Cells.
(A) Three replicates of Coomassie-stained SDS-gels with 4 different concentrations of cell culture and BSA proteinstandard (cell density of the cultures from top to bottom were: 1052, 1052 and 969 nl total cell volume*ml-1).(B) Three replicates of standard curves plotting BSA protein amounts versus band area calculated by the Quantity One 1-DAnalysis Software after background subtraction.(C) Three replicates of RbcL amounts per µl culture used for the calculation of RbcL amount after background subtraction.The µg RbcL/µl cell culture were then transformed to RbcL concentration [µM] considering the measured cell volume perml culture, the volume of the chloroplast, the absolute weight of RbcL and the Avogadro constant.
replicate 1
replicate 2
replicate 2 replicate 2
replicate 3
replicate 3 replicate 3
R² = 0.9964
0
0.2
0.4
0.6
0.8
1
0 50 100 150 200
Rb
cL a
mo
un
t [µ
g]
culture amount [µl]
replicate 1 replicate 1
R² = 0.9985
0
100
200
300
400
500
600
700
800
0 0.5 1 1.5
Ban
d a
rea
[mm
2]
BSA protein amount [µg]
R² = 0.9817
0
0.2
0.4
0.6
0.8
1
0 50 100 150 200
Rb
cL a
mo
un
t [µ
g]
culture amount [µl]
R² = 0.9967
0
200
400
600
800
1000
1200
0 0.2 0.4 0.6 0.8 1 1.2
Ban
d a
rea
[mm
2]
BSA protein amount [µg]
R² = 0.9979
0
0.2
0.4
0 20 40 60 80 100
Rb
cL a
mo
un
t [µ
g]Culture amount [µl]

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
1
10
100
1000
10000
0.0001 0.001 0.01 0.1 1 10 100 1000
Sub
stra
te [
S]
Substrate per binding site
3PGA
S7P
RuBP
SBP
DHAP
F6P
FBP
R5P
Xu5P
Ru5P
GAP
E4P
BPGA
2
1
23
4
4
5
5
5
6
7
7
7
7
7 9
8
6
10
1011 9
0
20
40
60
80
100
120
0.0001 0.001 0.01 0.1 1 10 100 1000
Satu
rati
on
[%
]
Substrate per binding site
RuBP
3PGA
BPGA
GAP
DHAP
FBP
SBP
S7P
E4P
R5P
Ru5P
2
1
7
73
45 555
6a
6b
8a
8b
1110
1 Rubisco2 PGK3 GAPDH4 TPI5 FBA + SBA6 FBPase
a reducedb oxidized
7 TRK8 SBPase
a reducedb oxidized
9 RPE10 RPI11 PRK
A
B
Supplemental Figure 20. Substrate per Binding Sites versus Substrate Concentrations (A) and Enzyme Saturation (B) ofCBC Enzymes.
Substrates of CBC reactions were measured via LC-MS/MS and are shown in Figure 6. Binding sites of CBC enzymes werecalculates based on proteomics data via the emPAI as described in text, Supplemental Table 1 and Methods.(A) Substrate per binding site of CBC reactions plotted against the substrate level before the light intensity was increasedfrom 41 to 145 µmol photons*m-2*s-1 (time point zero).(B) Substrate per binding site of CBC reactions plotted against the degree of saturation according to the saturation curvesshown in Figure 11, before the light intensity was increased from 41 to 145 µmol photons*m-2*s-1 (time point zero). For asimilar plot 20 min after the light shift, see Figure 13.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Figure 21. Percentage Change for Enzyme-substrate Pairs of the CBC between 20 min and 480 min Afterthe Light Shift Applied to C. reinhardtii CC-1690 Cells.
-120
-60
0
60
120
-15 -10 -5 0 5 10 15
% c
han
ge o
f su
bst
rate
leve
l b
etw
een
20
min
an
d 4
80
min
aft
er li
ght
shif
t
% change of protein abundance between 20 min and 480 min after light shift
all reactionsreactions with GAP as substrateallosterically regulated enzymes
R2=0.0115p-value=0.715
Rubisco (RuBP)
GAPDH (BPGA)
SBPase (SBP)
PRK (Ru5P)
RPI (R5P)
FBPase (FBP)
TPI (GAP)TRK (GAP)FBA (GAP)
TRK (F6P)
FBA (DHAP)
TRK (S7P)

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Substrate [µM] Substrate per binding site
Enzyme name Binding site [µM] Substratebefore light
shift20 min after
light shiftbefore light
shift20 min after
light shift
Rubisco 304.5 ± 32.6 RuBP 1057.5 5189.2 3.5 17.0
PGK 477.2 ± 54.5 3PGA 5261.3 7447.1 11.0 15.6
BPGA 2.8 4.0 0.0 0.0
GAPDH 651.5 ± 189.0 BPGA 2.8 4.0 0.0 0.0
TPI 44.3 ± 10.6 GAP 13.8 32.1 0.3 0.7
DHAP 509.4 1098.2 11.5 24.8
FBAa
658.5 ± 99.7 GAP 13.8 32.1 0.0 0.0
DHAP 509.4 1098.2 0.8 1.7
FBP 306.1 257.2 0.5 0.4
FBPase 121.0 ± 13.2 FBP 306.1 257.2 2.5 2.1
TRK 232.2 ± 47.0 F6P 498.9 1255.0 2.1 5.4
GAP 13.8 32.1 0.1 0.1
SBAa
658.5 ± 99.7 E4P 13.3 39.7 0.0 0.1
DHAP 509.4 1098,2 0.8 1.7
SBP 591.0 919.2 0.9 1.4
SBPase 149.6 ± 29.8 SBP 591.0 919.2 3.9 6.1
TRK 232.2 ± 47.0 S7P 1450.6 6048.2 6.2 26.0
GAP 13.8 32.1 0.1 0.1
R5P 76.1 133.7 0.3 0.6
Xu5P 43.7 85.1 0.2 0.4
RPE 87.7 ± 39.8 Xu5P 43.7 85,1 0.5 1.0
Ru5P 29.1 56.8 0.3 0.6
RPI 68.1 ± 14.8 R5P 76.1 133.7 1.1 2.0
Ru5P 29.1 56.8 0.4 0.8
PRK 451.3 ± 93.8 Ru5P 29.1 56.8 0.1 0.1
Supplemental Table 1. Binding Site Concentration of CBC Enzymes Based on emPAI Analysis of Proteomics Data.
Binding site concentrations at the lower light intensity were calculated by the emPAI and normalized to measuredamounts of Rubisco (for details see text and Methods). For comparison, substrate data of time point zero at the lowerlight intensity and 20 min after the light shift are shown in column four and five, respectively. In column six and seven theratio between substrate and binding site was calculated for time point zero and 20 min after the light shift, respectively.aOf the aldolase encoding genes, FBA1 and FBA3 were suggested to be localized in the chloroplast and FBA3 transcriptwas shown to be higher expressed in C. reinhardtii strain CC-4249 (Blaby et al., 2013). In this study, based on the amountof protein calculated via the emPAI-value, FBA3 protein was found to be >30-fold more abundant than FBA1. The bindingsite concentration for the aldolase reaction (FBA and SBA, two reactions catalysed by the same enzyme) is the sum of thecalculated abundance of FBA1 and FBA3.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Table 2. The Rate Equations of the CBC Reactions.
Rate equations according to Methods and Fridlyand and Scheibe (1999) used for calculation of substrate saturation curves(shown in Figure 11) together with the Km and KI values information of Supplemental Table 3 and metabolite amountsshown in Figure 6 and Supplemental Dataset 1.
Enzyme Rate equation
Rubisco 𝑣1 =𝑅𝑢𝐵𝑃 ∗𝑊𝐶∗min 1,
𝑅𝑢𝐵𝑃
𝐸𝑡
𝑅𝑢𝐵𝑃 +𝐾𝑚13 1+3𝑃𝐺𝐴
𝐾𝐼11+𝐹𝐵𝑃
𝐾𝐼12+𝑆𝐵𝑃
𝐾𝐼13+
𝑃𝑖𝐾𝐼14
+𝑁𝐴𝐷𝑃𝐻
𝐾𝐼15
𝑊𝐶 = 𝑉𝐶𝑚𝑎𝑥 ∗𝐶𝑂2
𝐶𝑂2 +𝐾𝑚11 1+𝑂2
𝐾𝑚12
PGK 𝑣2
𝑉𝑚𝑎𝑥=
3𝑃𝐺𝐴 ∗ 𝐴𝑇𝑃 −𝐵𝑃𝐺𝐴 ∗ 𝐴𝐷𝑃
𝐾𝐸2
𝐾𝑚21∗𝐾𝑚22∗ 1+3𝑃𝐺𝐴
𝐾𝑚71+
𝐴𝑇𝑃
𝐾𝑚72+𝐵𝑃𝐺𝐴
𝐾𝑚73+𝐴𝐷𝑃
𝐾𝑚74+3𝑃𝐺𝐴 ∗ 𝐴𝑇𝑃
𝐾𝑚71∗𝐾𝑚72+𝐵𝑃𝐺𝐴 ∗ 𝐴𝐷𝑃
𝐾𝑚73∗𝐾𝑚74
GAPDH 𝑣3𝑉𝑚𝑎𝑥
=𝐵𝑃𝐺𝐴 ∗ [𝑁𝐴𝐷𝑃𝐻]
𝐵𝑃𝐺𝐴 + 𝐾𝑚31 ∗ ([𝑁𝐴𝐷𝑃𝐻 + 𝐾𝑚32)
TPI 𝑣4
𝑉𝑚𝑎𝑥=
𝐺𝐴𝑃 −𝐷𝐻𝐴𝑃
𝐾𝐸4
𝐾𝑚41∗𝐾𝑚42∗ 1+𝐺𝐴𝑃
𝐾𝑚41+𝐷𝐻𝐴𝑃
𝐾𝑚42
FBA 𝑣5
𝑉𝑚𝑎𝑥=
𝐺𝐴𝑃 ∗ 𝐷𝐻𝐴𝑃 −𝐹𝐵𝑃
𝐾𝐸5
𝐾𝑚51∗𝐾𝑚52∗ 1+𝐺𝐴𝑃
𝐾𝑚51+𝐷𝐻𝐴𝑃
𝐾𝑚52+𝐺𝐴𝑃 ∗ 𝐷𝐻𝐴𝑃
𝐾𝑚51∗𝐾𝑚52+
𝐹𝐵𝑃
𝐾𝑚53
FBPase𝑣6
𝑉𝑚𝑎𝑥=
𝐹𝐵𝑃
𝐹𝐵𝑃 +𝐾𝑚61∗ 1+𝐹6𝑃
𝐾𝐼61+
𝑃𝑖𝐾𝐼62
SBA 𝑣7
𝑉𝑚𝑎𝑥=
𝐸4𝑃 ∗ 𝐷𝐻𝐴𝑃 −𝑆𝐵𝑃
𝐾𝐸7
𝐾𝑚71∗𝐾𝑚72∗ 1+𝐸4𝑃
𝐾𝑚71+𝐷𝐻𝐴𝑃
𝐾𝑚72+𝐸4𝑃 ∗ 𝐷𝐻𝐴𝑃
𝐾𝑚71∗𝐾𝑚72+
𝑆𝐵𝑃
𝐾𝑚73
SBPase𝑣8
𝑉𝑚𝑎𝑥=
𝑆𝐵𝑃
𝑆𝐵𝑃 +𝐾𝑚81∗ 1+𝑃𝑖
𝐾𝐼81
TRK 𝑣9
𝑉𝑚𝑎𝑥=
𝑆7𝑃 ∗ 𝐺𝐴𝑃 −𝑅5𝑃 ∗ 𝑋𝑢5𝑃
𝐾𝐸9
𝐾𝑚91∗𝐾𝑚92∗ 1+𝑆7𝑃
𝐾𝑚91+
𝐺𝐴𝑃
𝐾𝑚92+
𝑅5𝑃
𝐾𝑚93+𝑋𝑢5𝑃
𝐾𝑚94+
𝑆7𝑃 ∗ 𝐺𝐴𝑃
𝐾𝑚91∗𝐾𝑚92+𝑅5𝑃 ∗ 𝑋𝑢5𝑃
𝐾𝑚93∗𝐾𝑚94
RPI 𝑣10
𝑉𝑚𝑎𝑥=
𝑅5𝑃 −𝑅𝑢5𝑃
𝐾𝐸10
𝐾𝑚101∗ 1+𝑅5𝑃
𝐾𝑚101+
𝑅𝑢5𝑃
𝐾𝑚102
PRK𝑣11
𝑉𝑚𝑎𝑥=
𝑅𝑢5𝑃 ∗ 𝐴𝑇𝑃
𝑅𝑢5𝑃 +𝐾𝑚111∗ 1+3𝑃𝐺𝐴
𝐾𝐼111+𝑅𝑢𝐵𝑃
𝐾𝐼112+
𝑃𝑖𝐾𝐼113
∗ 𝐴𝑇𝑃 +𝐾𝑚112∗ 1+𝐴𝐷𝑃
𝐾𝐼114

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Table 3. Km and KI Values of Substrates and Inhibitors from CBC Enzymes.
Km and KI from literature that were used for calculation of substrate saturation curves (shown in Figure 11) together withthe rate equations given in Supplemental Table 2 and metabolite amounts shown in Figure 6 or Supplemental Dataset 1.
Enzyme EC Number ReactionSubstrate or
inhibitorParameter Value [mM] Organism Reference
Rubisco 4.1.1.39 RuBP+CO2→3PGA+3PGA CO2 Km11 0.033 Chlamydomonas reinhardtii Spreitzer et al. (1995)
O2 Km12 0.381 Chlamydomonas reinhardtii Spreitzer et al. (1995)
RuBP Km13 0.011 Chlamydomonas reinhardtii Spreitzer et al. (1995)
3PGA KI11 0.84 Spinacia oleaceae Badger and Lorimer (1981)
FBP KI12 0.04 Spinacia oleaceae Badger and Lorimer (1981)
SBP KI13 0.075 Spinacia oleaceae Badger and Lorimer (1981)
Pi KI14 0.9 Spinacia oleaceae Badger and Lorimer (1981)
NADPH KI15 0.07 Spinacia oleaceae Badger and Lorimer (1981)
PGK 2.7.2.3 3PGA+ATP↔BPGA+ADP 3PGA Km21 1.72 Spinacia oleaceae Köpke-Secundo et al. (1990)
ATP Km22 0.39 Spinacia oleaceae Köpke-Secundo et al. (1990)
BPGA Km23 0.004 Spinacia oleaceae Trost et al. (1993)
ADP Km24 0.27 Homo sapiens Varga et al. (2008)
KE2 7.6*10-4 Estimated Zhu et al. (2007)
GAPDH 1.2.1.13 BPGA+NADPH→GAP+NADP++Pi 3PGA Km31 0.015 Spinacia oleaceae Sparla et al. (2005)
ATP Km32 0.018 Chlamydomonas reinhardtii Graciet et al. (2003)
TPI 5.3.1.1 GAP↔DHAP GAP Km41 0.68 Spinacia oleaceae Harris and Koniger (1997)
DHAP Km42 2.5 Spinacia oleaceae Harris and Koniger (1997)
KE4 22.2 Chlorella pyrenoidosa Bassham and Krause (1969)
FBA 4.1.2.13 GAP+DHAP↔FBP GAP Km51 0.04 Spinacia oleaceae Iwaki et al. (1991)
DHAP Km52 0.45 Spinacia oleaceae Iwaki et al. (1991)
FBP Km53 0.0091 Pisum sativum Schnarrenberger et al. (1989)
KE5 7.1 Chlorella pyrenoidosa Bassham and Krause (1969)
FBPase 3.1.3.11 FBP→F6P+Pi FBP Km61(red) 0.06 Spinacia oleaceae Cadet and Meunier (1988)
FBP Km61(ox) 0.13 Spinacia oleaceae Cadet and Meunier (1988)
F6P KI61 0.7 Spinacia oleaceae Gardemann et al. (1986)
Pi KI62 12 Pisum sativum Charles and Halliwell (1981)
SBA 4.1.2.13 E4P+DHAP↔SBP E4P Km71 0.2 Estimated Zhu et al. (2007)
DHAP Km72 0.45 Spinacia oleaceae Iwaki et al. (1991)
SBP Km73 0.006 Daucus carota Moorhead and Planxton (1990)
SBPase 3.1.3.37 SBP→S7P+Pi SBP Km81(red) 0.05 Spinacia oleaceae Cadet and Meunier (1988)SBP Km81(ox) 0.18 Spinacia oleaceae Cadet and Meunier (1988)
Pi KI81 12000 Triticum aestivum Woodrow et al. (1983)
TRK 2.2.1.1 S7P+GAP↔R5P+Ru5P S7P Km91 0.46 Dictyostelium discoideum Albe (1991)
GAP Km92 0.072 Dictyostelium discoideum Albe (1991)
R5P Km93 0.33 Spinacia oleaceae Teige et al. (1998)
Xu5P Km94 0.067 Spinacia oleaceae Teige et al. (1998)
KE9 0.847 Chlorella pyrenoidosa Bassham and Krause (1969)
RPI 5.3.1.6 R5P↔Ru5P R5P Km101 0.63 Spinacia oleaceae Jung et al. (2000)
Ru5P Km102 0.66 Pisum sativum Skrukrud et al. (1991)
KE10 0.4 Chlorella pyrenoidosa Bassham and Krause (1969)
PRK 2.1.7.19 Ru5P+ATP→RuBP+ADP Ru5P Km111 0.056 Chlamydomonas reinhardtii Roesler et al. (1990)
ATP Km112 0.059 Spinacia oleaceae Gardemann et al. (1983)
3PGA KI111 2 Spinacia oleaceae Gardemann et al. (1983)
RuBP KI112 0.7 Spinacia oleaceae Gardemann et al. (1983)
Pi KI113 4 Spinacia oleaceae Gardemann et al. (1983)
ADP KI1140.04 Spinacia oleaceae Gardemann et al. (1983)

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Enzyme nameMethod A:
binding site [µM]Method B:
binding site [µM]
Rubisco 1006 ± 108 45.2 ± 4.8
PGK 1577 ± 180 70.8 ± 8.1
GAPDH 2153 ± 625 96.7 ± 28.0
TPI 146 ± 35 6.6 ± 1.6
aFBA 2176 ± 330 97.7 ± 14.8
FBPase 400 ± 44 18.0 ± 2.0
TRK 767 ± 155 34.4 ± 7.0
aSBA 2176 ± 330 97.7 ± 14.8
SBPase 494 ± 99 22.2 ± 4.4
TRK 767 ± 155 34.4 ± 7.0
RPE 290 ± 132 13.0 ± 5.9
RPI 225 ± 49 10.1 ± 2.2
PRK 1491 ± 310 67.0 ± 13.9
Supplemental Table 4. Binding Site Concentration of CBC Enzymes Based on emPAI Analysis of Proteomics Data with TwoDifferent Methods.
Method A: The relative abundance calculated by the emPAI (see Methods) was normed on total protein.Method B: The relative abundance calculated by the emPAI (see Methods) was normed on total protein after correctingthe latter for the fraction of the total annotated C. reinhardtii proteins that were detected in our analysis (n = 4) (i.e. aftermultiplying total protein by number of detected proteins/number of annotated proteins= 767/17038). aSee legend ofSupplemental Table 1.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Metabolite levels[µM C atoms]c T0.5 [s] Calculated duration
to change pool size[s]eMetabolite Stoichiometry low light 5min higher light low light 5min higher light
RuBP 1a 1648.58 5485.39 2.44 3.56 11.11
3PGA 2a 7030.66 7715.67 20.80 10.01 3.97
GAP 2a 27.72 60.38 0.08 0.08 0.19
DHAP 2a 1021.13 2003.29 3.02 2.60 5.69
FBP 0.33a 636.34 470.63 0.31 0.10 0.16
F6P 0.33a 2320.71 4712.93 1.14 1.02 2.31
E4P 0.33a 53.07d 137.85d 0.03 0.03 0.08
SBP 0.33a 1289.95 1641.70 0.64 0.35 0.34
S7P 0.33a 3166.06 10157.40 1.56 2.20 6.75
R5P 0.33a 118.66 198.95 0.06 0.04 0.08
Xu5P+Ru5P 1a 113.54 193.38 0.17 0.13 0.23
G1P 0.01b 501.38 549.95 0.07 0.12 2.76
G6P 0.01b 7515.67 10029.70 1.11 0.65 -2.05
ADPG 0.034b 169.97 431.23 0.01 0.03 0.37
UDPG 0.034b 571.50 687.22 0.59 0.31 0.23
Supplemental Table 5. Estimated Turnover Rates of CBC Intermediates in C. reinhardtii CC-1690 Cells.
Half-times (T0.5) of metabolites and time to change pool sizes from low to higher light intensity were calculated based oncarboxylation rates derived from measured net photosynthesis rates (338 and 771 µmol*s-1*L cell volume-1 respectively)and considering pathway stoichiometry and number of C atoms of the metabolites (Arrivault et al., 2009).aCBC intermediates: the number of turnovers per molecule of RuBP synthesized.bEnd-product synthesis: number of turnovers per molecule of CO2 fixed, taking into account the number of carbonmolecules in the metabolite and the stoichiometry of the synthesis pathway.cThe measured concentration of metabolite [µM] multiplied by the number of C atoms gives the concentration of Catoms in the corresponding metabolite pool.dCalculation of E4P assuming equilibrium of the TRK-catalysed reaction (F6P + GAP ↔ E4P + Xu5P). For details seeSupplemental Dataset 2.eDuration of CO2 fixation at the new light intensity that is required to produce the measured change in the size of thatmetabolite pool in the first 5 min after the increase in light intensity. The sum for all CBC metabolites is 32 s.

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental Table 6. Carbon Sequestration into Metabolites after Transfer to Higher Light.
For details of calculation see text and Methods.
% C sequestered into starch and metabolite in the time intervals
0-5min 5-10min 10-20min 20-40min 40-60min 60-120min 120-240min 240-480min
2OG -0.03 0.07 0.04 -0.04 0.01 0.00 0.00 0.00
Aconitate 0.00 0.01 0.00 0.00 0.00 0.00 0.00 0.00
ADPG 0.54 0.29 -0.11 -0.03 0.01 -0.02 -0.01 0.00
Aspartate -0.26 0.09 -0.22 -0.38 -0.02 -0.04 0.09 -0.02
Citrate 0.01 1.07 0.10 -0.10 -0.37 0.12 -0.02 -0.03
DHAP 0.76 0.05 0.05 -0.01 0.00 0.00 0.00 0.00
F6P 1.85 0.15 0.36 -0.14 -0.03 0.03 0.02 0.02
FBP -0.13 -0.02 0.03 0.01 0.00 0.00 0.00 0.00
G1P 0.04 0.05 0.00 -0.02 0.00 0.01 0.00 0.01
G3P 0.14 0.04 0.01 -0.01 -0.02 0.00 0.00 0.00
G6P 1.94 0.63 -0.01 -0.44 -0.09 0.04 0.02 0.11
GAP 0.03 0.01 0.00 0.00 0.00 0.00 0.00 0.00
Glutamate -0.80 2.61 1.11 -0.23 0.26 0.17 -0.01 0.06
Glycerate 0.01 -0.04 0.01 0.00 -0.01 0.00 0.00 0.00
Isocitrate 0.03 0.03 -0.02 0.02 0.00 0.01 0.00 0.00
Malate 4.52 1.27 0.54 -0.46 0.19 -0.01 0.22 0.00
PGA 0.53 2.82 -0.55 0.11 -0.09 0.11 -0.19 -0.01
R5P 0.06 0.01 0.00 -0.01 0.00 0.00 0.00 0.00
RuBP 2.96 1.82 0.10 0.31 -0.15 0.05 -0.08 -0.07
S7P 5.40 1.38 0.48 -0.31 -0.07 0.10 0.01 0.00
SBP 0.27 0.08 0.10 0.00 -0.07 0.01 0.00 -0.01
Shikimate 0.28 -0.21 0.14 -0.29 -0.01 -0.03 0.02 -0.02
Succinate -0.27 -0.19 0.22 0.09 -0.07 -0.03 0.03 -0.02
UDPG 0.22 -0.03 0.00 -0.05 0.02 0.00 0.00 0.00
Xu5P + Ru5P 0.06 0.01 0.01 -0.01 0.00 0.00 0.00 0.00
Starch 59.70 55.79 38.89 24.97 10.11 -4.93 10.63 3.08

Supplemental Data. Mettler et al. Plant Cell (2014) 10.1105/tpc.114.124537
Supplemental References
Albe K. R. (1991). Partial Purification and Kinetic Characterization of Transaldolase from Dictyostelium discoideum.Exp. Myc. 1.5: 255-262.Badger, M.R., and Lorimer, G.H. (1981). Interaction of sugar phosphates with the catalytic site of ribulose-1,5-bisphosphate carboxylase. Biochemistry 20: 2219-2225.Cadet, F., and Meunier, J.C. (1988). pH and kinetic studies of chloroplast sedoheptulose-1,7-bisphosphatase fromspinach (Spinacia oleracea). Biochem. J. 253: 249-254.Charles, S.A., and Halliwell, B. (1981). Light activation of fructose bisphosphatase in photosynthetically competentpea chloroplasts. Biochem. J. 200: 357-363.Chen, Y.R., Larimer, F.W., Serpersu, E.H., and Hartman, F.C. (1999). Identification of a catalytic aspartyl residue ofD-ribulose 5-phosphate 3-epimerase by site-directed mutagenesis. J. Biol. Chem. 274: 2132-2136.Graciet, E., Lebreton, S., Camadro, J.M., and Gontero, B. (2003). Characterization of native and recombinant A4glyceraldehyde 3-phosphate dehydrogenase. Kinetic evidence for conformation changes upon association with thesmall protein CP12. FEBS 270: 129-136.Iwaki T., Wadano A., Yokota A. Himeno M. (1991). Aldolase - an important enzyme in controlling the ribulose 1,5-bisphosphate regeneration rate in photosynthesis. Plant Cell Physiol. 32: 1083-1091.Jung, C.H., Hartman, F.C., Lu, T.Y.S., and Larimer, F.W. (2000). D-ribose-5-phosphate isomerase from spinach:Heterologous overexpression, purification, characterization, and site-directed mutagenesis of the recombinantenzyme. Arch. Biochem. Biophys. 373, 409-417.Kopke-Secundo, E., Molnar, I., and Schnarrenberger, C. (1990). Isolation and characterization of the cytosolic andchloroplastic 3-phosphoglycerate kinase from spinach leaves. Plant Physiol. 93: 40-47.Moorhead, G.B., and Plaxton, W.C. (1990). Purification and characterization of cytosolic aldolase from carrotstorage root. Biochem. J. 269: 133-139.Roesler, K.R., and Ogren, W.L. (1990). Primary structure of Chlamydomonas reinhardtii ribulose 1,5-bisphosphatecarboxylase/oxygenase activase and evidence for a single polypeptide. Plant Physiol 94, 1837-1841.Schnarrenberger, C.; Kruger, I (1986). Distinction between cytosol and chloroplast fructose-bisphosphatealdolases from pea, wheat, and corn leaves. Plant Physiol. 80: 301-304.Skrukrud, C.L., Gordon, I.M., Dorwin, S., Yuan, X.H., Johansson, G., and Anderson, L.E. (1991). Purification andcharacterization of pea chloroplastic phosphoriboisomerase. Plant Physiol. 97: 730-735.Sparla, F., Zaffagnini, M., Wedel, N., Scheibe, R., Pupillo, P., and Trost, P. (2005). Regulation of photosyntheticGAPDH dissected by mutants. Plant Physiol. 138: 2210-2219.Spreitzer, R.J., Thow, G., and Zhu, G. (1995). Pseudoreversion substitution at large-subunit residue 54 influencesthe CO2/O2 specificity of chloroplast ribulose-bisphosphate carboxylase/oxygenase. Plant Physiol. 109: 681-685.Teige, M., Melzer, M., and Suss, K.H. (1998). Purification, properties and in situ localization of the amphibolicenzymes D-ribulose 5-phosphate 3-epimerase and transketolase from spinach chloroplasts. Eur. J. Biochem. 252:237-244.Thimm, O., Blasing, O., Gibon, Y., Nagel, A., Meyer, S., Kruger, P., Selbig, J., Muller, L.A., Rhee, S.Y., and Stitt, M.(2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways andother biological processes. Plant J. 37: 914-939.Trost P., Scagliarini S., Valenti V., Pupilio P. (1993). Activation of spinach chloroplast glyceraldehyde 3-phosphatedehydrogenase: effect of glycerate 1,3-bisphosphate. Planta 190:321-326.Varga, A., Szabo, J., Flachner, B., Roy, B., Konarev, P., Svergun, D., Zavodszky, P., Perigaud, C., Barman, T.,Lionne, C., and Vas, M. (2008). Interaction of human 3-phosphoglycerate kinase with L-ADP, the mirror image ofD-ADP. Biochem. Biophys. Res. Commun. 366: 994-1000.Woodrow, I.E., Murphy, D.J., and Walker, D.A. (1983). Regulation of photosynthetic carbon metabolism. Theeffect of inorganic phosphate on stromal sedoheptulose-1,7-bisphosphatase. Eur. J. Biochem. 132: 121-123.