Data. Bedewitz et al. Cell (2014) 10.1105/tpc.114.130534...Supplemental Data. Bedewitz et al. Plant...
Transcript of Data. Bedewitz et al. Cell (2014) 10.1105/tpc.114.130534...Supplemental Data. Bedewitz et al. Plant...

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Figure 1. Assignment of Gene Ontology terms to A. belladonna transcripts. Plant GO-Slim terms were assigned for each GO each category.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Figure 2. Number of A. belladonna unigenes in the different clusters as identified by OrthoMCL.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Ab-ArAT3 1 -MEICSTKS--RWNFQGKEEAKKASACTIRSYLNTLNESINKN-D-------------TRIVIPLSHGDPSGFPNFRTTK Solyc-ArAT3 1 -MEVSSK---WRWNFEGKEEAKTASACTIRSYLNTLNKNINKNDN-------------TKSVIPLSHGDPSGFPSFRTSK Ab-ArAT4 1 -MEKGND-------HETKEITEIPKNITIRSVLGLLMANTDGD-E-------------KKKVISLGIGDPTAYSCFRTTD Solyc-ArAt4 1 -MGSQTN----YLGENYTKELETPTNITIKKILGLLMGIIDGD-DE-----------RKREVISLGIGDPTAHSCFHTTN Ab-ArAT5 1 -MENIGSVTNISFGENYTKQVETPTNITIKGILGLLMDNIEGDEG-------------KREVISLGIGDPTAHSCFHTTD Solyc-ArAt5 1 -MENIGSQTNISF---QTKQVESFTNITIKGILGLLMDTIEEGKD-------------KKEVISLGIGDPTAHSCFHTTD At5g53970 1 -MENGAT---------------TTSTITIKGILSLLMESITTEEDEG-----------GKRVISLGMGDPTLYSCFRTTQ Solyc-ArAt1 1 MEGNLVG------AINNQVEMETPNNITIKGILGLLMANTEAT-D-------------VKKVISLGMGDPTLYSCFHSPD Ab-ArAT1 1 METNGVKVKANLSDGVNNQEIETPNNITIKGILGLLMANVETASDEENGCNNLKNNKSKKKVISLGMGDPTAYSCFHSPA Solyc-ArAt6 1 -MENKLS----KWGFNNGDSKEK-SLFSIRSVLETIMENLKED-D-------------ERVMIHLGRGDPSSIPCFRTSP At5g36160 1 MGENGAK----RWNFGANEVVERSNSLTIRDYLNTLINCLDGG-D-------------VRPVIPLGHGDPSPFPSFRTDQ Ps-TyrAT 1 -MEKGGK----KWIIRGNDKLKVGTENTIRGLLEVMNSNLNVN-D-------------ERPIIPLGHGDPSPFTCFRTTH Ab-ArAT2 1 -MENGTTTS--KWNFKENEKLVSVSGLTVRSVLNKLMSGVDPA-D-------------TRPVIPLGHGDPSAFPCFRTTP Solyc-ArAt2 1 -MENGTTTGMRRWNFKENEKLVSVSDLTVRSVLNKLRCCVDPA-D-------------TRPTIPLGHGDPSAFPCFLTTT Ab-ArAT3 64 LSEDALVDALQSGKYNGYSQKSAILSATRSIAEYLSRDYPYELSPNDVHVTAGAKQAIDVLITALAVPGANILLPRPGYP Solyc-ArAT3 64 VSEDALVDALQSAKYNGYCGNPDIFHASRSIAEYLSRDYPYKLSPNDVLVTAGAKHGIDVLITALAVPGANILLPRPGYP Ab-ArAT4 59 AAKQVVADSLLSDKYNGYPPAVGLPRTREAIADYLSRDIPYKVSANDVYITAGCTQAIEIVLTILARPGANILLPRPGFS Solyc-ArAt4 64 EAKEAVVESLLIDKFNGYSPSAGLPTTRKAIASYLSRDLCYELSQEDVYITVGCTQAIEMALSFLAKPNSNILLPRPGFP Ab-ArAT5 67 AAKEAVVESLLLDKFNGYSPTSGLPAARKAIANYLSRDLPYDLSPEDVYVTAGCTQAIEIAVSILAKPNSNILLPRPGFP Solyc-ArAt5 64 AAKEAVVESLLLDKFNGYSPSNGLAIARKAIANYLSRDLPYDLSAEDVYVTSGCTQAIEIAVSILAKPNSNILLPRPGFP At5g53970 54 VSLQAVSDSLLSNKFHGYSPTVGLPQARRAIAEYLSRDLPYKLSQDDVFITSGCTQAIDVALSMLARPRANILLPRPGFP Solyc-ArAt1 61 VAHDAVIESLTSHKFNGYSPTVGLPQTRKAIADYLSRELPEKLCADDVYITAGCTQAIELALSILARPGANILLPRPGFP Ab-ArAT1 81 VAQDAVLETLASNKFNGYAPTVGLPQTRKAIADYLSRDLPEKLCADDVYVTAGCTQAIEIALSILARPGSNILLPRPGFP Solyc-ArAt6 61 VAEDAIFGAVRSAKFNGYAPAAGIYPARRSIAEYLSHDLPHHVSPDDVFLTPGANQAIEVVMSVLARPGANILFPKPGYP At5g36160 63 AAVEAICDAVRSTKFNNYSSSSGVPVARKAVAEYLSSDLSYQISPNDVHITAGCVQAIEILISALAIPGANILLPRPTYP Ps-TyrAT 62 IVDDALNTAIQSAKFNSYPPPAGIPTARRSIAEHLSRDLPYKLSTEDVFLTSGCRQAIEIITTVLACPGGNILIPKPGYP Ab-ArAT2 64 VAEDAISDAVRSAKFNGYSSTVGILPARRAVAEYLSQDLPYKLSPDDVYLTIGCNQAIEILLNALARPNANILLPTPGFP Solyc-ArAt2 66 IAEDAISDAVRSAKFNGYSSTVGILPARRAVAEYLSQDLPYKLSPDDIYLTSGCGQAIEILLNALARPNANILLPSPGFP Ab-ArAT3 144 AYEALATFNGLEMRHYDLLPEQDWEVDINGLEALADDRTVAMVVINPGNPCGNVYKREHLKKIAEAARKLGMLVISDEAY Solyc-ArAT3 144 AYEALATFNRLEMRHYDLLPEQDWEVDIDGLEALADDKTVAMVVINPGNPCGNVYEREHLKKIAEAASKLGMLVISDEAY Ab-ArAT4 139 IYSLCAAFRNLEIRYYDLLPEKGWEVDLSAVESLANENTVGLVVINPGNPCANVYSHQHLEKIAQTAKRLQTVVVADEVY Solyc-ArAt4 144 TYALCASFRNIEVRYYDLLPQNNWQLNLEAIEDLVDQNTIAIVIINPGNPCGNVYSYDHLKKIAETAKKVKTLIIADEVY Ab-ArAT5 147 IYALCAAFRNVEVRYYDLVPEKNWEVNLKAIEDLADQNTVAIVVINPGNPCGNVYSYEHLEKIAETAKRVKTLIIEDEVY Solyc-ArAt5 144 TYVLCAAFRNVQVRYYDLLPQNNWQVDLKSIEDLADQNTIGIVVINPGNPCGNVYSYDHLEKIAETAKKVKTLIIADEVY At5g53970 134 IYELCAKFRHLEVRYVDLLPENGWEIDLDAVEALADENTVALVVINPGNPCGNVYSYQHLMKIAESAKKLGFLVIADEVY Solyc-ArAt1 141 IYALCAAFRNIEVRYFDLIPDKGWEVDLNAVEALADHNTIGIVVINPGNPCGNVYSYQHLQQIAETAKKLRTIVIADEVY Ab-ArAT1 161 IYALCAAFRHIEVRYFDLVPGKGWEVDLNAVEALADRNTIAIVVINPGNPCGNVYSYQHLQEIAETAKKLRTIVIADEVY Solyc-ArAt6 141 FYDARAACSNLEVRHFDLLPEKGWKVDLESVEALVDDHTIAIVIINPGNPCGNVFTSEHLQEIAETAKKLGILVIADEVY At5g36160 143 MYDSRAAFCQLEVRYFDLLPENGWDVDLDGVEALADDKTVAILVINPCNPCGNVFSRQHLQKIAETACKLGILVIADEVY Ps-TyrAT 142 HYDACAVFHNLEVRHFDLLPEKAWEVDLDAVEALADENTVAIVIINPGNPCGNVYTYEHLKKVAETAKRLGIPVIADEVY Ab-ArAT2 144 YYETWAGFSHLEMRHFNLLPEKEWEVDLNMVESLADENTVAIVIINPGNPCGNVYTDQHLKKVAEMARKHGILVIADEVY Solyc-ArAt2 146 YYEAWGGFTQMEMRHFNLLPEKEWEVDLNAVESLADENTVAMVIINPGNPCGNVYSEEHLKKVAETARKLGILVISDEVY Ab-ArAT3 224 GHLVFGRNPFVPMGVFGEIAPILTIGSISKRWMVPGWRFGWIVICDLNGILQKHGVVESVKNYLNISVVPPTLTLGAIPR Solyc-ArAT3 224 GHLVFSSNPFVPMGVYGEIAPILTIGSISKRWMVPGWRFGWIVMCDPNGILQKHGVAESIKNYLNIGVGPPTFTLGAIPR Ab-ArAT4 219 GHLAFDENPFVPMAAFSSLVPVLTLGSLSKRWLVPGWRLGWFVINDPNAIFVNPKFMERIRNYCDICRGPATFIQAAVPA Solyc-ArAt4 224 GHLAFGETPFTPMGLFSSITPILTLGSLSKRWLIPGWRLGWFVINDPNCIFKSPKIIERINNYCDLCASPTTFIQAAVPK Ab-ArAT5 227 GHLAFGENPFISMGLFSSIAPILTLGSLSKRWLIPGWRLGWFVVNDPNCIFKSPKIVERIKKYCDICGGPATFIQAAVPK Solyc-ArAt5 224 GHLAFGENPFISMGLFSSITPILTLGSLSKRWLIPGWRLGWFVINDPNCIFKSPKIIERIKKYCDIGGGPATFIQAAVPK At5g53970 214 GHLAFGSKPFVPMGVFGSIVPVLTLGSLSKRWIVPGWRLGWFVTTDPSGSFKDPKIIERFKKYFDILGGPATFIQAAVPT Solyc-ArAt1 221 GHLAFGANPFVPMGIFGDIAPVLTLGSLSKRWLVPGWRLGWLVTNDPNGTFKNPKFVERIKKYCDICGGPATFIQAAVPR Ab-ArAT1 241 GHLAFGAKPFVPMGVFGAIAPVLTLGSLSKRWLVPGWRLGWLVTNDPNGTFKNPKFVERIKKYCDICGGPATFIQAAVPR Solyc-ArAt6 221 SHLCFGSKPFVPMGVFGSITPILTLGSISKRWVVPGWRLGWIATIDPSGVLKKSGIAECLQNYLEYSANPATIVQGAVPH At5g36160 223 DHFAFGDKPFVSMAEFAELVPVIVLGAISKRWFVPGWRLGWMVTLDPHGIMKDSGFVQTLINVVNMSTDPATFIQGAMPD Ps-TyrAT 222 AHLIFGSNPFVPMGVFGSTVPIFTLGSISKRWIVPGLRLGWLVITDPSGFLKDTEIVSLIKQCLNMSTSPACVIQGALPQ Ab-ArAT2 224 AHLTFGSKPFVPMGIFGSIAPVITLGSISKRWIVPGWRLGWLVTNDPNGILKEHGVIDSITGYLNISSDPATFIQGAIPQ Solyc-ArAt2 226 AHLAFGSKPFVPMGIFGSIAPVVTLGSISKRWIVPGWRLGWLVTNDPNGILKEHGDIDSIMGYLNISTDPATFIQGAIPQ

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Ab-ArAT3 304 ILVETPEDFYSNTINLLRKAADICYAGLREIPCFT-PYKPQGSMFLMAKLNMSLLEGINNDKEFCTRLAREESVVVLPGE Solyc-ArAT3 304 FLVETPEDFYSNIINLLRKAADICYAGLREIPCFT-PYKSQGSMFLMAKLNMSLLEGIDNDMEFCTRLAREESAIVLPGE Ab-ArAT4 299 IIEQTTETFYQKTISMLKHTSNLCYEKIKEIPWLICPCKPQGAMFLMVKLNCPQLEDISDDLDFCFKLAKEESVIILPGV Solyc-ArAt4 304 IIENTKEDFFKNTLKMLKKNSDICYEKILEIPCLNCPYKPQGSMVVMVNLNIILLQNIIDDMDFCFKLAKEEYVILLPGT Ab-ArAT5 307 IIESTKEDFFRNTLKMLKKNSDMCYEKILEIPCINCPYKSQGSMVVMVKFNSALLQDISDDIDFCFKLAKEESVILLPGT Solyc-ArAt5 304 IIESTKEDFFKNTLKMLKKNSDICYEKIQEISCINCPYRSQGSMVLMVKLNLTLLKDISDDIDFCFKLAKEESVLLLPGT At5g53970 294 ILEQTDESFFKKTLNSLKNSSDICCDWIKEIPCIDSSHRPEGSMAMMVKLNLSLLEDVSDDIDFCFKLAREESVILLPGT Solyc-ArAt1 301 IIQQTEDVFFRKTVNLLKWTADICCDKIKEIPCISCPYKPEGSMAVMVKLHLPLMIDISDDIDFCFKLAKEESVILLPGL Ab-ArAT1 321 IIQQTEDVFFRKTVNLLKWTADICCEKIKEIPCISCPHKPEGSMAVMVKLHLSLLEDISDDIDFCFKLAKEESIILLPGL Solyc-ArAt6 301 ILEKTTKEFFSHINNVLKEAVDAFYSKVQEIPCFICPDKPEGAMSMMIKLNLSLLEDINDDMDFCTKLAHEESVLILPGK At5g36160 303 IIGNTKEEFFSSKLEMVKKCAEICYEELMKIPCITCPCKPEGSMFTMVKLNFSLLEDISDDLDFCSKLAKEESMIILPGQ Ps-TyrAT 302 ILENTKEDFFENIISLLCQAIDICYEEIKEIACITLLPRPEGSMFLMVKLNTVLLEDISDDMDFCFKLAKEESVIVLPGA Ab-ArAT2 304 ILQKTKDDFFSKIVNMLRDDADIVYDRIKDIPCITCPSKPEGSMFVMVKLHLDLLEDIEDDLDFCYKLAKEESLITLPGV Solyc-ArAt2 306 ILHETKDDFFSKIVNMLREDADICYERIKDIPCITCPSKPQGSMFLMVQLHLNLLEDIEDDLDFCAKLAKEESLIILPGV Ab-ArAT3 383 ALGLKSWVRVTFAVEISALEDGLGRIKAFCFRNDKQQ-------- Solyc-ArAT3 383 ALGLKNWVRVTFAVEISALEDGLSRIKAFCFRNAKQQ-------- Ab-ArAT4 379 AVGMRNWVRITFAVEPSSLEEALARVKCFCLNHTKQENGHTPSID Solyc-ArAt4 384 AVGLKNWIRITFAVESSFLEEGLKRLKSFCLRHSKPQNLIN---- Ab-ArAT5 387 AVGLKNWLRITFAVEPSFLEEGLRRLKTFCLRHSNPKTLS----- Solyc-ArAt5 384 AVGLKNWLRITFAVDVSFLEEGLSRLKSFCLRHSNTKIMI----- At5g53970 374 AVGLKNWLRITFAADATSIEEAFKRIKCFYLRHAKTQYPTI---- Solyc-ArAt1 381 AVGLKNWIRITFAADPASLEEALGRLKSFCQRHSYQQNDHH---- Ab-ArAT1 401 AVGLKNWIRITFAADPSSLEEALGRLKSFCQRHSCQQNGHH---- Solyc-ArAt6 381 IVGLKNWLRLTFAMEPAILEEALERIKSFYLRHSKVKGL------ At5g36160 383 AVGLKNWLRITFAVELELLIEGFSRLKNFTERHSKNQP------- Ps-TyrAT 382 VLGLENWLRITFSIDLASLKDGLQRMKMFCRRHAKQQ-------- Ab-ArAT2 384 AVGLKNWLRITFACEPSSLEDGFQRLNAFYQRHAKKQ-------- Solyc-ArAt2 386 AVGLKNWLRITFACEPSYLEDGFQRLNAFYKRHAKKQ--------
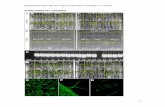
Supplemental Figure 3. Amino Acid Alignment of Ab-ArATs and Selected Homologs. A full-length amino acid alignment of the Ab-ArATs and related homologs from tomato (Solanum lycopersicum), Arabidopsis thaliana and poppy (Papaver somniferum) was generated using MUSCLE (Edgar, 2004). BoxShade was utilized to highlight conserved amino acids as indicated by the shaded squares. The catalytic lysine is shown in red and conserved residues required for binding of the pyridoxal 5´-phosphate cofactor are shown in gold.
Edgar, R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792-1797.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
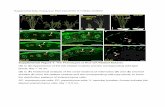
Supplemental Figure 4. The Utility of Virus-Induced Gene Silencing for Functional Analysis of Alkaloid Biosynthesis in A.belladonna.
(A) and (B) Silencing of PHYTOENE DESATURASE (PDS) in A.belladonna compared to TRV2-empty vector controls. Note the characteristic photo-bleaching of PDS-silenced lines.
(C) Silencing of ACTIN in A.belladonna compared to TRV2-empty vector controls. Note the characteristic stunting of growth in the ACTIN-silenced lines.
(D) through (I) The impact of silencing PUTRESCINE METHYLTRANSFERASE (PMT) and HYOSCYAMINE-6-HYDROXYLASE (H6H) on tropane alkaloid and selected intermediate compound levels in A.belladonna. Two separate constructs designated H6H1 and H6H2 that were designed to target separate regions of the H6H gene were utilized. Plants infected with the TRV2 empty vector were used as controls. Data are presented as the mean of at least n = 11 ± SE. Asterisks denote significant differences from the TRV2 empty vector controls (*, p < 0.05; **, p < 0.01; ***, p < 0.001) as determined by Student’s t test. Experimental procedures are detailed in the METHODS.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Figure 5. The Impact of Silencing Additional ArATs on Tropane Alkaloid Biosynthesis in A. belladonna.
(A) Abundance of scopolamine in TRV2 empty vector and TRV2:Ab-ArAT VIGS lines. Data are presented as the mean n = 12 ± SE. Asterisks denote significant differences (* = p < 0.05) as determined by Student’s t-test. (B) Relative expression level of the targeted Ab-ArATs in TRV2 empty vector control lines and TRV2:Ab-ArAT VIGS lines. Data were obtained by qRT-PCR and presented as the mean of six biological and three technical replicates with the expression level of the target in TRV2 empty vector control lines set to 1 ± SE. Asterisks denote significant differences (** = p < 0.01; *** = p < 0.001) as determined by Student’s t-test.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Figure 6. Potential Forward and Reverse Reactions Catalyzed by ArATs.
The forward reaction consists of an aromatic amino donor (group A) and a 2-oxo acid amino acceptor (group B), with amino group transfer resulting in the conjugate 2-oxo acid ( group C) and conjugate amino acid (group D). The reverse direction would involve an amino acid from group D as the amino donor, and a 2-oxo acid from group C as the amino acceptor, yielding an aromatic amino acid (group A) and the conjugate 2-oxo acid (group B). Reactions proceed in both directions until equilibrium is reached.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Figure 7. Kinetic analysis of the transaminase reaction catalyzed by 6xHis-Ab-ArAT4. Experimental details are provided in the METHODS section of the manuscript. Substrate and co-substrate combinations are as follows: (A) 4-Hydroxyphenylpyruvate with 10 mM L-Phenylalanine. (B) L-Phenylalanine with 7 mM 4-Hydroxyphenylpyruvate. (C) 2-Oxoglutarate with 10 mM L-Phenylalanine. (D) L-Phenylalanine with 25 mM 2-Oxoglutarate. (E) Phenylpyruvic Acid with 4 mM L-Tyrosine. (F) L-Tyrosine with 1 mM Phenylpyruvic Acid. (G) Oxaloacetate with 10 mM L-Phenylalanine. (H) L-Tryptophan with 7 mM 4-Hydroxyphenylpyruvate. (I) Pyruvic Acid with 10 mM L-Phenylalanine. Data are presented as the mean of three experiments, ± SE.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Figure 8. Physical Properties of the Ab-ArAT4 Enzyme. (A) The influence of pH on Ab-ArAT4 activity. (B) Determination of the T1/2 (temperature at which 50% of activity is lost) of Ab-ArAT4. T1/2 is defined as the temperature at which 50% of enzyme activity is lost. Experimental details are provided in the METHODS section of the manuscript. In all assays, enzyme activity was determined by LC-MS/MS using a standard curve of L-Tyr with data presented as the mean of 3 replicates ± SE.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 1. Proteins used for phylogenetic analysis.a
aPhylogenetic analysis of these proteins is presented in Figure 4.
Species Gene name Identifier Source Atropa belladonna Ab-ArAT1 KC954703 www.ncbi.nlm.nih.gov/
Ab-ArAT2 KC954704 www.ncbi.nlm.nih.gov/
Ab-ArAT3 KC954705 www.ncbi.nlm.nih.gov/
Ab-ArAT4 KC954706 www.ncbi.nlm.nih.gov/
Ab-ArAT5 KC954707 www.ncbi.nlm.nih.gov/ Solanum lycopersicum Solyc-ArAT1 Solyc10g007110 http://solgenomics.net/
Solyc-ArAT2 Solyc07g053720 http://solgenomics.net/
Solyc-ArAT3 Solyc07g053710 http://solgenomics.net/
Solyc-ArAT4 Solyc12g096240 http://solgenomics.net/
Solyc-ArAT5 Solyc12g088000 http://solgenomics.net/
Solyc-ArAT5 Solyc10g008200 http://solgenomics.net/ Solanum pennellii Sopen-ArAT1 ADZ24702 www.ncbi.nlm.nih.gov/
Sopen-ArAT2 ADZ24703 www.ncbi.nlm.nih.gov/
Solanum tuberosum Sotub-ArAT1 Sotub10g008150 http://potatogenomics.plantbiology.msu.edu
subgroup phurea Sotub-ArAT2 PGSC0003DMT400068957 http://potatogenomics.plantbiology.msu.edu
Sotub-ArAT3 Sotub07g021530 http://potatogenomics.plantbiology.msu.edu
Sotub-ArAT4 Sotub12g028590 http://potatogenomics.plantbiology.msu.edu
Sotub-ArAT5 Sotub12g026080 http://potatogenomics.plantbiology.msu.edu
Sotub-ArAT6 Sotub10g008680 http://potatogenomics.plantbiology.msu.edu
Nicotiana benthamiana Niben-ArAT1a NbS00042707g0006 http://solgenomics.net/
Niben-ArAT1b NbS00045248g0003 http://solgenomics.net/
Niben-ArAT2a NbS00000301g0008 http://solgenomics.net/
Niben-ArAT2b NbS00009959g0008 http://solgenomics.net/
Niben-ArAT2c NbS00008749g0006 http://solgenomics.net/
Niben-ArAT3a NbS00046710g0015 http://solgenomics.net/
Niben-ArAT3b NbS00000144g0005 http://solgenomics.net/
Petunia hybrida Ph-PPY-AT AHA62827 www.ncbi.nlm.nih.gov/
Arabidopsis thaliana AT5G53970 AT5G53970 www.arabidopsis.org
AT5G36150 AT5G36150 www.arabidopsis.org
Papaver somniferum Ps-Tyr-AT GU370929 www.ncbi.nlm.nih.gov/
Perilla frutescens Pf-ArAT ADO17550 www.ncbi.nlm.nih.gov/
Solenostemon scutellarioides Ss-ArAT CAD30341 www.ncbi.nlm.nih.gov/
Cucumis melo Cm-ArAT1 ADC45389 www.ncbi.nlm.nih.gov/
Salvia miltiorrhiza Sm-ArAT ABC60050 www.ncbi.nlm.nih.gov/
Bruguiera gymnorrhiza Bg-ArAT ADD54646 www.ncbi.nlm.nih.gov/

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 2. Estimates of the Average Evolutionary Divergence of Solanaceae ArATs.
Ab-ArAT4 Clade A Clade B Clade C Clade D Clade E
Ab-ArAT4 0
Clade A 0.35 (0.008)
0.16 (0.021)
Clade B 0.33 (0.002)
0.33 (0.003)
0.10 (0.009)
Clade C 0.49 (0.006)
0.47 (0.003)
0.45 (0.003)
0.13 (0.024)
Clade D 0.51 (0.003)
0.51 (0.003)
0.51 (0.002)
0.44 (0.005)
0.11 (0.020)
Clade E 0.46 (0.004)
0.46 (0.002)
0.42 (0.003)
0.39 (0.003)
0.43 (0.002)
0.16 (0.013)
The p-distance (1-amino acid identity) from averaging over all sequence pairs between and within clades A through E and in comparison to Ab-ArAT4 (Figure 4) is shown. Standard error estimates are shown in parentheses. Positions containing gaps and missing data were eliminated. Analyses were performed using MEGA5 (Tamura et al., 2011).
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731-2739.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 3. Amino acid levels (pmoles mg dwt-1) in TRV2 empty vector control lines and TRV2:Ab-ArAT4 VIGS lines. Data presented are the mean of 17 or 21 biological replicates ± SE. P-values were determined using the Student’s t-test. Amino Acid TRV2 TRV2:Ab-ArAT4 P-value Alanine 5635 ± 489 5563 ± 651 0.93 Arginine 51 ± 3 73 ± 3.6 <0.001a
Asparagine 2847 ± 185 3347 ± 233 0.11 Aspartic Acid 6099 ± 377 6154 ± 540 0.94 Cysteine 13 ± 1 12 ± 1 0.72 Glutamine 60416 ± 3982 53538 ± 2864 0.16 Glutamic Acid 96093 ± 4395 91043 ± 3886 0.39 Glycine 15559 ± 1030 13242 ± 676 0.06 Histidine 758 ± 29 721 ± 28 0.38 Isoleucine and Leucine 525 ± 22 533 ± 27 0.83 Lysine 632 ± 21 760 ± 46 0.02 Methionine 1294 ± 85 1124 ± 52 0.08 Phenylalanine 2023 ± 77 1940 ± 67 0.42 Proline 4171 ± 480 3789 ± 555 0.67 Serine 1895 ± 123 1628 ± 89 0.11 Threonine 6306 ± 194 6674 ± 362 0.41 Tryptophan 797 ± 55 693 ± 40 0.13 Tyrosine 1005 ± 55 893 ± 32 0.07 Valine 1670 ± 80 1659 ± 67 0.91
a Significant differences at a p-value of 0.05 are highlighted in bold.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 4. Oligonucleotide primers used in this study
Primer Sequence Use Ab-PDS VF 5´-CGACGACAAGACCCTGGAGATGCTTTCGTGTTTGCCA-3´ VIGS construct assembly Ab-PDS VR 5´-GAGGAGAAGAGCCCTGGAGTTTTGACAACATGATACT-3´ VIGS construct assembly Ab-ACT VF 5´-CGACGACAAGACCCTTGGAGATGGTGTCAGCCACACA-3´ VIGS construct assembly Ab-ACT VR 5´-GAGGAGAAGAGCCCTGACACTGTATTTCCTCTCAGGC-3´ VIGS construct assembly Ab-PMT VF 5´-CGACGACAAGACCCTGATTGTTCATCTCCCACTTGGTTC-3´ VIGS construct assembly Ab-PMT VR 5´-GAGGAGAAGAGCCCTTGCTGGACCAATAGGATCAGAAGA3´ VIGS construct assembly Ab-H6H1 VF 5´-CGACGACAAGACCCTGTGCCTATCATTGATCTCCAACA-3´ VIGS construct assembly Ab-H6H1 VR 5´-GAGGAGAAGAGCCCTAGTCAGCATCATCTGAATTTGGC-3´ VIGS construct assembly Ab-H6H2 VF 5´-CGACGACAAGACCCTGCCAGCTAAATTTGAGCTTCCA-3´ VIGS construct assembly Ab-H6H2 VR 5´-GAGGAGAAGAGCCCTGGTTCAACAGCAATCCATTTTG-3´ VIGS construct assembly Ab-ArAT1 VF 5´-CGACGACAAGACCCTTTCTACTTAGTAAGCAACCCGG-3´ VIGS construct assembly Ab-ArAT1 VR 5´-GAGGAGAAGAGCCCTAGACTCAAAATCAATATTAATT-3´ VIGS construct assembly Ab-ArAT2 VF 5´-CGACGACAAGACCCTGCTACTCTGAGAAGCAACCTT-3´ VIGS construct assembly Ab-ArAT2 VR 5´-GAGGAGAAGAGCCCTTGAATATGCACATTTCACAAA-3´ VIGS construct assembly Ab-ArAT3 VF 5´-CGACGACAAGACCCTAATGATAAGGAATTCTGTACCA-3´ VIGS construct assembly Ab-ArAT3 VR 5´-GAGGAGAAGAGCCCTAAATTTCCAACAGGCTTAACAGC-3´ VIGS construct assembly Ab-ArAT4 VF 5´-CGACGACAAGACCCTGCAGTGGAGCCATCTTCCCTGG-3´ VIGS construct assembly Ab-ArAT4 VR 5´-GAGGAGAAGAGCCCTCTTCATATTACATAGGAATACTAAT-3´ VIGS construct assembly Ab-ArAT5 VF 5´-CGACGACAAGACCCTTGAGTGAAAATATGGAGAATATAGG-3´ VIGS construct assembly Ab-ArAT5 VR 5´-GAGGAGAAGAGCCCTGGTGATAAATCATATGGAAGATC-3´ VIGS construct assembly Ab-ArAT6 VF 5´-CGACGACAAGACCCTGGTTGATCCCAGTGGCGTCCTTAAG-3´ VIGS construct assembly Ab-ArAT6 VR 5´-GAGGAGAAGAGCCCTCAACCAATTCTTTAGTCCCACAATC-3´ VIGS construct assembly Ab-ArAT4 ENT-F 5´-CACCATGGAGAAAGGGAATGATCATGA-3´ Expression construct assembly Ab-ArAT4 ENT STOP-R 5´-TTAGTCGATGGAGGGAGTGTGTCCA-3´ Expression construct assembly Ab-EF1-F 5´-CCCAAGAGGCCATCAGACA-3´ qRT-PCR Ab-EF1-R 5´-ACCAATCTTGTAAACGTCCTGAAGT-3´ qRT-PCR Ab-ArAT1 QF 5´-CGGAGTTAACAACCAAGAAATTGA-3´ qRT-PCR Ab-ArAT1 QR 5´-TTGGCCATTAACAATCCAAGAA-3´ qRT-PCR Ab-ArAT2 QF 5´-GGTGTTGCTGTGGGACTCAA-3´ qRT-PCR Ab-ArAT2 QR 5´-TCCAGAGATGATGGCTCACATG-3´ qRT-PCR Ab-ArAT3 QF 5´-ACCTACTGAGAAAAGCAGCAGACA-3´ qRT-PCR Ab-ArAT3 QR 5´-TTGTATGGAGTAAAGCACGGAATC-3´ qRT-PCR Ab-ArAT4 QF 5´-CCTCTGCTTTCTCTATTTTGCATTG-3´ qRT-PCR Ab-ArAT4 QR 5´-TTTGTTTCATGATCATTCCCTTTCT-3´ qRT-PCR Ab-ArAT5 QF 5´-ATTCAGCCCTCCTCCAAGATATAA-3´ qRT-PCR Ab-ArAT5 QR 5´-GGATTCCTCCTTGGCAAGCT-3´ qRT-PCR Ab-ArAT6 QF 5´-GGAGCCAGCCATTCTTGAAG-3´ qRT-PCR Ab-ArAT6 QR 5´-TTTAGCTACTTGTATGCCTCAAGCA-3´ qRT-PCR

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 5. HPLC mobile phase gradient utilized for LC-MS/MS Analyses of Alkaloids and Select Precursors in Positive Mode. Mobile phase A = 1mM aqueous perfluoroheptanoic acid. Mobile phase B = acetonitrile. See METHODS section of manuscript for details.
Time (min) Mobile phase A (%)
Mobile phase B (%)
0.00 99 1 0.75 99 1 1.33 75 25 2.70 60 40 3.70 40 60 3.71 5 95 4.33 5 95 4.34 99 1 5.00 99 1

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 6. Multiple Reaction Monitoring Parameters Utilized for LC-MS/MS Analyses of Tropane Alkaloids and Select Precursors in Positive Mode.
Compound Precursor ion >
product ion (m/z)
Cone voltage
(V)
Collision voltage
(V)
Retention time (min)
Putrescine 89 > 72 20 10 3.72 N-methylputrescine 103 > 72 50 10 3.78
Tropinone 140 > 98 28 22 2.77 Tropine 142 > 98 40 22 2.73
Hyoscyamine 290 > 124 46 22 3.76 Scopolamine 304 > 103 34 40 3.51

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 7. HPLC Mobile Phase Gradients Utilized for LC/MS/MS Analyses of Phenylpyruvic Acid and DL-3-Phenyllactic Acid using an Applied Biosystems MDS SCIEX 3200 QTRAP Mass Spectrometer. Mobile phase A = 0.15% aqueous formic acid. Mobile phase B = methanol. See METHODS section of manuscript for details.
Time (min) Mobile phase A (%)
Mobile phase B (%)
0.00 98 2 0.01 98 2 0.40 98 2 3.00 60 40 5.50 0 100 8.50 0 100 8.51 98 2
10.00 98 2

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 8. Multiple Reaction Monitoring Parameters Utilized for LC/MS/MS Measurements of Phenylpyruvic Acid and DL-3-Phenyllactic Acid on an Applied Biosystems MDS SCIEX 3200 QTRAP Mass Spectrometer.
Compound Precursor ion > product ion
(m/z)
Declustering potential (V)
Collision voltage (V)
Retention time (min)
Phenylpyruvic acid 163.05 > 91 -40 -20 4.44 DL-3-Phenyllactic acid
Propyl-4-Hydroxybenzoatea 165.05 > 103 179.06 > 92
-30 -40
-20 -20
4.85 6.04
a 10μM Propyl-4-Hydroxybenzoate was included as an internal standard.

Supplemental Data. Bedewitz et al. Plant Cell (2014) 10.1105/tpc.114.130534
Supplemental Table 9. HPLC Mobile Phase Gradients Utilized for LC-MS/MS Analyses of Phenylpyruvic Acid using a Waters Quattro Premier XE mass spectrometer. Mobile phase A = 0.1% aqueous formic acid. Mobile phase B = methanol. See METHODS section of manuscript for details.
Time (min) Mobile phase A (%)
Mobile phase B (%)
0.00 98 2 0.40 98 2 0.60 60 40 5.80 40 60 6.00 0 100 7.00 0 100 9.00 0 100 9.02 98 2
10.00 98 2