Supplemental Data. Peng et al. (2011). Plant Cell 10.1105 ... · 10/31/2011 · Peng et al....
Transcript of Supplemental Data. Peng et al. (2011). Plant Cell 10.1105 ... · 10/31/2011 · Peng et al....

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Figure 1. The genetic map position of the fcl1 locus on chromosome 6. (A) A
linkage map based on the recombination frequency analysis of 254 individuals from an F2
mapping population and twelve SSR markers initially identified to be co-segregated with the
fcl1 locus from a bulked segregant analysis. (B) A fine genetic map based on the recombinant
frequency analysis of 213 fcl1 mutant plants from an F2 mapping population of 787
individuals. The fcl1 locus was linked to the SSR marker AC141436-ssr4.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Figure 2. Confirmation of deletion junctions and expression analysis of genes
deleted in fcl1-1 and fcl1-2 alleles. (A) PCR amplification of deletion junctions in fcl1-1 (lanes
1, 2) and fcl1-2 (lanes 3, 4). A17, wild-type plants; MW, molecular weight ladders. (B) Reverse
transcription (RT)-PCR amplification of transcripts encoded by the twelve predicted ORFs
deleted in the fcl1-1 and fcl1-2 alleles in vegetative shoot apices of WT plants. A high level of
transcripts was detected for ORFs 1, 3, 4, 6, 11 and 12. Primer sequences used are listed in
Supplemental Table 4.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Figure 3. Allelic series tests of fcl1 mutants. (A-E) Four-week-old WT (cv.
Jemalong A17; A), fcl1-2 (B), WT (cv. R108; C), fcl1-3 (D) and an F1 plant from a cross
between fcl1-2 and fcl1-3 (E). (F) The Tnt1 insertion site in fcl1-3 and fcl1-4 mutants. Open
box, untranslated region; solid box, exon; horizontal line, intron and arrowhead, orientation of
the open reading frame. (G) PCR-based genotyping. Lane 1, PCR amplification of the deletion
junction in the fcl1-2 mutant. Lane 2, PCR amplification of the FCL1 gene in WT (cv. Jemlong
A17 or R108). Lane 3, PCR amplification of the Tnt1 insertion site in the fcl1-3 mutant. (H)
Compound leaf phenotype of wild-type (left) and the fcl1-4 mutant (right). (I) Reduced petiole
length of compound leaves from different nodes of the stem in fcl1-4 mutant. Shown were
mean ± s.d., n=3.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Figure 4. Compound leaf phenotypes of fcl1-4 mutant. (A) Three-month-old
fcl1-4 (-/-) homozygous plants (right) exhibited various compound leaf phenotypes, including
leaflet clustering and fusion. fcl1-4 (+/-) heterozygous plants (left) were used as a comparison,
showing wild-type-like trifoliolate compound leaves. (B) Close-up views of compound leaves
of fcl1-4 (+/-) heterozygous plants and fcl1-4 (-/-) homozygous plants as indicated.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Figure 5. Genetic complementation of fcl1-1. (A, B) Transgenic fcl1-1 mutant
harboring a 7.8kb genomic sequence of FCL1 (fcl1-1/FCL1), showing both leaflet fusion and
reduced petiole phenotypes of the mutant were rescued.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
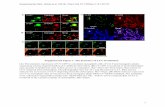
Supplemental Figure 6. Zinc finger domain transcription factor Medtr6g080720.1 did not
rescue fcl1-1 compound leaf defects. (A) Image of a stably transformed fcl1-1 plant. (B) RT-
PCR analysis of gene expression in transgenic fcl1-1 plants. Compared with wild-type
(Jemalong A17) and fcl1-1 mutant, the zinc finger transcription factor (ZnF) was not expressed
in the fcl1-1 mutant but was expressed in transgenic fcl1-1 mutant transformed with the WT
gene similarly to the WT plants. However, the expression of the ZnF gene did not rescue the
mutant compound leaf defects.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
1 aatgttttgagataaatagtatatatgagcagtgttgtagatgtggtatatcaaaggttt
61 ttatttcatctactcaaaaatagttatatggaatctaagATGAATAGTTTTGTTGTTGAT
M N S F V V D
121 CATGAGAAGTACAAACAAGAAGAAAATCAAGAAGAATTGGAATTACAACATAAAGAAGGT
H E K Y K Q E E N Q E E L E L Q H K E G
181 GGTGTTCATGAACATGATCATCAAGAGGAAAATGATGAGATTCTTAAGACAAGAATCTCT
G V H E H D H Q E E N D E I L K T R I S
241 AACCATCCTCTCTATGAACTTCTTGTTCAAGCTCACTTGGATTGTTTGAAGGTTGGTGAC
N H P L Y E L L V Q A H L D C L K V G D
301 ATTTCTAACTTGGAGATAGAGAAATCAGATAAAAAGCAAACACTGAAGAAACAAAACTTG
I S N L E I E K S D K K Q T L K K Q N L
361 GATATGTTAAGCCAATCTGAGCTTGATCTCTTCATGGAAGCATATTGTTTGGCACTTAGT
D M L S Q S E L D L F M E A Y C L A L S
421 AAGCTAAAAGAAGCAATGAAGGAACCACAACAGAATTCAATGGCTTTCATAAACAACATG
K L K E A M K E P Q Q N S M A F I N N M
481 CATTCACAACTTAGGGAACTAACTCAGGCAACTTCATCTTCTAGTGAACCAGATGCTACT
H S Q L R E L T Q A T S S S S E P D A T
541 ACATCTTCAAGTGAGTGCACATTCAGAAGAAATCCATCAATTTAGaggttgatgacttga
T S S S E C T F R R N P S I *
601 ttttcagtagtgtggttgtgcatgtaatatcctaagataggttaattaattaaggtttca
661 attattgtcaaattgtcatgtccatgtttgaaatgttattttatgtgaaattaatgggca
721 atgaatgttgttataaattaattaatctatagtatttccccttatctcattaattaattg
781 cagttgttatctac
Supplemental Figure 7. Full-length FCL1 cDNA and the deduced amino acid sequences.
Lower case letters indicate 5’- and 3’- untranslated regions (UTRs). Capital letters indicate the
coding region. Red and blue arrowheads indicate the Tnt1 insertion sites in fcl1-3 and fcl1-4
mutants, respectively. Red and cyan shaded letters indicate the KNOX1 and KNOX2 domains,
respectively. An asterisk indicates the stop codon.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplement Figure 8. Amino acid sequence alignments of FCL1 and its close homologs.
FCL1 amino acid sequences were used as seeds to BLAST search the NCBI GenBank. The
identified homologous sequences were manually examined to ascertain that full-length
sequences do not contain the homeodomain. Sequences were aligned using Clustal X.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
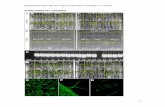
Supplemental Figure 9. Microarray-based in silico analysis of FCL1 tissue-specific
expression. Tissue-specific expression of FCL1 was analyzed using Affymetrix GeneChip
Medicago Genome Array-based Medicago Gene Expresion Atlas
(http://bioinfo.noble.org/gene-atlas/v2/).

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Figure 10. Quantitative RT-PCR analysis of KNOX2 (A) and KNOX6 (B)
expression in vegetative shoot buds and young leaves in wild-type and fcl1-1 mutant. Shown
were mean ± s.d., n=3.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Table 1. Segregation ratio of fcl1-2 mutants in F2 mapping populations
Population Genetic crosses wild type mutant total 𝑿𝟐 (P value)
1 fcl1-2 x Jemalong A20 175 79 254 5.045 (0.025)
2 fcl1-2 x Jemalong A20 399 134 533 0.006 (0.940)
Total fcl1-2 x Jemalong A20 574 213 787 0.179 (0.181)
𝑋2, Chi square based on 3:1 segregation ratio.

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128 Supplemental Table 2. Recombination frequency and genetic distance between the fcl1-2 locus and molecular markers on chromosome 6
Marker Recombination frequencies (± S.E.) Genetic distance (± S.E.) cM
DK321L 0.021 ± 0.007 2.114 ± 0.007
44D11L 0.005 ± 0.003 0.469 ± 0.003 005D01 0.005 ± 0.003 0.469 ± 0.003 AC141436-ssr4 0 ± 0 0 ± 0 002E12 0.014 ± 0.006 1.409 ± 0.006
002F12 0.021 ± 0.007 2.114 ± 0.007 18A5R 0.026 ± 0.007 2.585 ± 0.008 19O4L 0.026 ± 0.008 2.585 ± 0.008

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Table 3. Genotypes of 20 fcl1-2 mutant plants that carry recombination between fcl1
and closely linked markers
Marker
fc11-2 plant
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
DK321L B B B B B B B H B H H H H H B B H H H B
005D01 B B B B B B B B B B B B B B B B H B H B
44D11L B B B B B B B B B B B B B B B B H B H B
AC141436-ssr4 B B B B B B B B B B B B B B B B B B B B
002E 12 B B B H H B H B H B B B B B H H B B B B
002F 12 H H H H H B H B H B B B B B H H B B B B
18A5R H H H H H H H B H B B B B B H H B B B H
19O4L H H H H H H H B H B B B B B H H B B B H
H, heterozygous mutant marker; B, homozygous mutant marker

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Table 5. Arabidopsis thaliana homologs of deleted genes in fcl1 mutants
Deleted gene Encoded
protein
Homolog Annotation Other names
Medtr6g080670.1
(partial)
424 a.a. AT3G17810 dihydropyrimidine
dehydrogenase
PYD1;
MEB5.3;
PYRIMIDINE
1
Medtr6g080680.1 68 a.a. No homolog
Medtr6g080690.1 161 a.a. AT1G14760.2 Class M Knox protein KNATM-B
Medtr6g080700.1 357 a.a. AT2G02650.1 Putative non-LTR
retroelement reverse
transcriptase
Medtr6g080710.1 31 a.a. No homolog
Medtr6g080720.1 1179 a.a. AT1G10170.1 Zinc finger domain
transcription factor
ATNFXL1
Medtr6g080730.1 154 a.a. AT4G33420.1 Putative peroxidase
Medtr6g080740.1 73 a.a. AT4G33420.1 Putative peroxidase
Medtr6g080750.1 60 a.a. No homolog
Medtr6g080760.1 53 a.a. No homolog
Medtr6g080770.1 161 a.a. AT3G05540.1 Translationally
controlled tumor protein
TCTP
Medtr6g080780.1
(partial)
356 a.a. AT1G66200.1 glutamate-ammonia
ligase
ATGSR2

Supplemental Data. Peng et al. (2011). Plant Cell 10.1105/tpc.111.089128
Supplemental Table 6. Double mutant analyses of F2 plants
Genetic cross WT-like sgl1- like fcl1- like plant with novel phenotype
sgl1-1 x fcl1-1 308 69 101 32
Genetic cross WT-like fcl1-like palm1-like plant with novel phenotype
fcl1-2 x palm1-2 400 186 124 0