Supplemental Data. Gong et al. (2012). Plant Cell 10.1105 ... · 9/11/2012 · Plant Cell...
Transcript of Supplemental Data. Gong et al. (2012). Plant Cell 10.1105 ... · 9/11/2012 · Plant Cell...

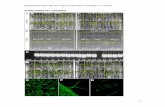
Supplemental Figure 1. Fine structure of the functional core of potato Cen6. The top track in each panel shows the sequence coordinates on the respective potato chromosome. The “annotated genes” track illustrates the positions of annotated genes. The green line in the middle track shows the number of sequence reads derived from ChIP with CENH3 antibodies in 100 bp windows. The vertical red bars represent the percentage of the 32 tissues in which the corresponding gene is expressed (FPKM >0) (1 representing expression in all 32 tissues). The bottom track shows the density of reads in 100 bp windows, adjusted by the length of mappable regions. The horizontal green bars in this track mark the sequencing gap/non-mappable regions. Each green bar region is assigned to an adjacent CENH3 subdomain. All CENH3 subdomains are shaded in yellow.
Supplemental Data. Gong et al. (2012). Plant Cell 10.1105/tpc.111.100511

Supplemental Figure 2. Fine structure of the functional core of potato Cen11. The top track in each panel shows the sequence coordinates on the respective potato chromosome. The “annotated genes” track illustrates the positions of annotated genes. The green line in the middle track shows the number of sequence reads derived from ChIP with CENH3 antibodies in 100 bp windows. The vertical red bars represent the percentage of the 32 tissues in which the corresponding gene is expressed (FPKM >0) (1 representing expression in all 32 tissues). The bottom track shows the density of reads in 100 bp windows, adjusted by the length of mappable regions. The horizontal green bars in this track mark the sequencing gap/non-mappable regions. Each green bar region is assigned to an adjacent CENH3 subdomain. All CENH3 subdomains are shaded in yellow.

Supplemental Figure 3. Fine structure of the functional core of potato Cen12. The top track in each panel shows the sequence coordinates on the respective potato chromosome. The “annotated genes” track illustrates the positions of annotated genes. The green line in the middle track shows the number of sequence reads derived from ChIP with CENH3 antibodies in 100 bp windows. The vertical red bars represent the percentage of the 32 tissues in which the corresponding gene is expressed (FPKM >0) (1 representing expression in all 32 tissues). The bottom track shows the density of reads in 100 bp windows, adjusted by the length of mappable regions. The horizontal green bars in this track mark the sequencing gap/non-mappable regions. Each green bar region is assigned to an adjacent CENH3 subdomain. All CENH3 subdomains are shaded in yellow.

Supplemental Figure 4. ChIP-qPCR confirmation of potato genes associated with CENH3 nucleosomes. Primers designed from two expressed genes, PGSC0003DMG400012074 (2074) and PGSC0003DMG400039086 (9086), and three silenced genes, PGSC0003DMG400012078 (2078), PGSC0003DMG400045728 (5728), and PGSC0003DMG400008748 (8748), were used in qPCR analysis. The actin-97 gene (PGSC0003DMG400027746), which is located outside of the centromeres, was used as a negative control. The actin-97 sequence showed no enrichment in both computer-based mapping and qPCR analysis. All five genes were mapped to centromeres and were highly enriched with CENH3 in computer-based analysis. Relative fold enrichment (RFE) is normalized with the actin-97 control, then calculated as 2-ΔCT (see Methods) and indicated by the height of the color-coded columns with standard error. ChIP-qPCR confirmed the significant enrichment of all five genes in ChIPed DNA sample as compared to the negative control. The “-1, -2, -3, -5, -6” suffixes on X-axis represent different primer pairs for the same gene.

Supplemental Figure 5. Proportion of repeat families in WGS-454 and CENH3 ChIP-seq data. Red dots represent 454 clusters.

Supplemental Figure 6. Representative fiber-FISH images from six centromere-specific satellite repeats. The fiber-FISH signals were digitally straightened. Each bar represents 50 µm, which spans approximately 150 kb of DNA .

Supplemental Table 1. Measurements of fiber-FISH signals from six centromeric repeats[A conversion rate of 3.21 kb/µm (Cheng et al. 2002) was used to convert the measurements]
Repeat and Fibers Fiber Length (µm) Fiber Length (kb)St3-58 fibers1 502.2376 1612.1832 477.3534 1532.3043 459.043 1473.5284 439.6233 1411.1915 415.8535 1334.896 402.0071 1290.443
Average 449.353 µm 1442.423 kb
St49 fibers1 736.0371 2362.6792 660.5066 2120.2263 621.0397 1993.5374 591.6801 1899.2935 591.6314 1899.1376 578.5979 1857.299
Average 629.92 µm 2011.029 kb
St24 fibers1 1275.562 4094.5552 1546.391 4963.9153 1216.949 3906.4084 1364.087 4378.7195 1293.657 4152.638
Average 1339.329 4299.247
St3-238 fibers*1 1319.578 4235.8452 1275.946 4095.7873 837.6238 2688.7724 807.0844 2590.7415 763.9987 2452.4366 531.4964 1706.103
St18 fibers*1 2397.52 7696.032 2177.56 6989.963 1275.07 4092.964 1218.61 3911.75

5 1073.97 3447.466 1064 3415.447 821.89 2638.268 821.15 2635.919 733.13 2353.35
St57 fibers1 245.071 786.6792 326.843 1049.1673 250.240 803.2704 291.661 936.2315 252.279 809.8166 240.468 771.9037 330.391 1060.5558 284.689 913.8509 288.463 925.96510 236.066 757.77311 314.576 1009.79012 313.159 1005.24213 242.147 777.29114 334.550 1073.90515 250.101 802.82316 272.400 874.40317 277.511 890.81118 236.165 758.08819 236.637 759.60320 372.428 1195.49521 331.761 1064.95422 276.954 889.02323 237.591 762.66724 359.391 1153.646
Average 283.398 µm 909.706 kb*Lengths of the fibers are high variable, the short fibers most likely represent broken fibers.

Supplemental Table 2. PCR primers used for cloning potato satellite repeats Repeat Primer Sequence [5'->3'] St3-58 CL3c58-R1 GTACATAAACATGCTCACCAAGTAAG
CL3c58-F2 TGTTGGTTGGGATATAAATTATGC
St3-238 CL3c238-R1 AGGCTTAACCGACATAGACCATAC
CL3c238-F2 ATGGGATGTGCACTCTAGTATGGTC
St3-294 CL3c294-F2 TTAGGACAATCTCTCACCTTGTG
CL3c294-R1 ATTTACAAAAATTGTGGATGCTATG
St18 CL18c4-F3 ACTTGGGAACATTCGTCCTC
CL18c4-R3 CCCTCAAAATGAATTAGGATGAAC
St24 CL24c27-F3 CGAGCTGACTTCCACATCG
CL24c27-R3 TGAAGCACACCTTCAGTTGC
St49 CL49c3-F3 AACTAACTAGAGATGCTGGGACAAG
CL49c3-R3 GTTACCCGCTTGACGTAACAC
St57 CL57c12-F3 CCTCATATGTATGCCCTCATTG
CL57c12-R3 TGTATTGGTTCCTACGCTCTACC

Supplemental Table 3. Primer set used to analyze putative active genes located in the CENH3 domain
Expected amplicon length Primer Primer sequence Target gene
cDNA gDNA
Function of the target gene
2074 F ACGTGGTGGAGACTGAACCT PGSC0003DMG400012074 165 165 Hypothetical protein
2074 R CTCCAACCTCACCCTTTCCT
9086_1 F AAATTGGTGCTGATGGGGTA PGSC0003DMG400039086 207 1661 TRNA-dihydrouridine synthase
9086_1 R CGCACGTATGCGTCAGTAGT
9086_2 F CGAACTGTTGAATGGGGATT 185 1638
9086_2 R TGACCTGGAAGCTGAGAACC
3879 F ACACCAATGACCCAACAACA PGSC0003DMG400043879 352 1008 Hypothetical protein
3879 R GAAACAGCAGCAAAGGAACC
actin-97 F AGTATGACGAATCTGGTCCTTCTATTG St-actin 97 203 203 Actin
actin-97 R ACCCAACAATCAACTCTGCCCTCTC
urease F TTGGACATTGACGGATCAGA PGSC0003DMG400001246 196 1388 Urease
urease R TTACGCCTCCTTTGATCACC