Supplemental InformationAS ON-KL Edited · Supplemental Data. Stepanova et al. Plant Cell. (2011)....
Transcript of Supplemental InformationAS ON-KL Edited · Supplemental Data. Stepanova et al. Plant Cell. (2011)....
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
1
Supplemental Information
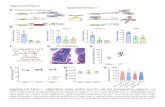
Supplemental Figure 1. Loss- and gain-of-function analysis of the putative PDC gene family in Arabidopsis. (A) Phylogenetic tree of the PDC and PDC-like proteins in Arabidopsis. Protein alignment and the phylogenetic tree were made with the Geneious package (www.geneious.com). The following parameters were used for the protein alignment: cost matrix Blosum 62, gap opening penalty 5.0, gap extension penalty 3.0, and refinement iterations 2. For the phylogenetic tree construction, the Jukes-Cantor genetic distance model and the Neighbor-Joining tree building method were utilized. Consensus tree was generated by resampling using the Bootstrap method with a sample
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
2
size of 1000 reiterations and a support threshold of 50%. (B) Schematic representation of the PDC gene structure and positions of the T-DNA insertions in the mutant alleles characterized in this study. (C) Origin and molecular nature of the mutant alleles described. (D) Triple response phenotypes of loss-of-function (pdc1-10, pdc2-10, pdc3-10, and pdc4-11) and gain-of-function mutants of PDC1 through 4. Seedlings of the indicated genotypes were grown for three days in the dark in AT plates with or without the ethylene precursor ACC.
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
3
Supplemental Figure 2. Simultaneous activation of the CYP79B2/B3 and YUC
pathways in the sur2 YUC1ox lines leads to additive phenotypic defects. (A)
Quantification of the hypocotyl lengths in three-day-old etiolated seedlings of Col, sur2,
Col YUC1ox, and sur2 YUC1ox. The first four bars represent average lengths (±SD) of at
least 20 seedlings of the indicated genotypes. The “Additive” bar shows the expected
hypocotyl length of sur2 YUC1ox under a simple additive model. (B-D) Phenotypes of
the same lines grown in plates for three days in the dark (B), three days in the dark
followed by four days in constant light (C), or for three weeks in soil under 16h light/ 8h
dark cycle (D). Images of the plants marked with asterisks are the same as in Figure 2.
Representative plants are displayed.
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
4
Supplemental Figure 3. TAA1:YUC1 can suppress the flower defects of yuc1/2/4/6,
but not of wei8-2 tar2-1. Inflorescence stems of soil-grown Col, wei8-2 tar2-1, and
yuc1/2/4/6 with or without the TAA1:YUC1 transgene are shown.
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
5
Supplemental Figure 4. Functional TAA1 and TARs are not required for iaaM-
mediated auxin biosynthesis. In the upper panels, representative Col, wei8-2 tar2-1, and
wei8-2 tar2-1 TAA1:iaaM plants grown for four weeks in soil under a 16h light/ 8h dark
cycle are shown. In the lower panels, representative inflorescence stems of the
corresponding genotypes are displayed.
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
6
Supplemental Figure 5. Exogenous IAA can restore some but not all of the wei8
tar2 mutant phenotypes. Col and wei8-2 tar2-1 were grown in plates containing 0, 0.01,
0.1, 0.5, or 1 µM IAA. A) Representative dark-grown three-day-old seedlings are shown.
B) Representative root meristems of seedlings germinated for three days in the dark
followed by seven additional days of growth in constant light are displayed.
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
7
Supplemental Figure 6. Representative chromatograms of free IAA and IAA
catabolites/conjugates. Samples from a Col-‐0 wild-‐type extract are compared to a
standard mixture, both injected onto a Kinetex C18 HPLC column. Separation of
unlabeled/labeled analytes: oxIAA/13C6-‐oxIAA (A), IAAsp/13C6-‐IAAsp (B),
IAGlu/13C6-‐IAGlu (C), and IAA/13C6-‐IAA (D).
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
8
Supplemental Table 1. Primers used to genotype the different mutants employed in this study
Line Locus Origin Reference mutant fragment WT fragment
wei8-1 At1g70560 Act. Tag T-DNA Stepanova et al, 2008 DWLB1 + 70560-R1 70560-F8 + 70560-R1
wei8-2 At1g70560 EMS Stepanova et al, 2008 70560-F9 + 70560-R1 N/A
tar1-1 At1g23320 GABI566A11-2 Stepanova et al, 2008 DWLB1 + TAR1-R1C TAR1-F1C + TAR1-R1C
tar2-1 At4g24670 Salk_021258 Stepanova et al, 2008 24670-F1 + JMLB1 24670-F1 + 24670-R1
tar2-2 At4g24670 Salk_137800 Stepanova et al, 2008 24670-F1 + JMLB1 24670-F1 + 24670-R1
tar2-3 At4g24670 Sail_351_F01 Stepanova et al, 2008 LB3SAIL + 24670-R1 24670-F1 + 24670-R1
sur2 At4g31500 Salk_028573 Stepanova et al, 2005 CYP83B1-F + JMLBa CYP83B1-F1 + CYP83b1-R1
cyp79b2 At4g39950 Pooled Salk collection Zhao et al, 2002 CYP79B2-F +
JMLBa CYP79B2-F + CYP79B2-R
cyp79b3 At2g22330 Pooled Salk collection Zhao et al, 2002 JMLB1 +
CYP79B3-R CYP79B3-F + CYP79B3-R
yuc1 At4g32540 Salk_106293 Cheng et al, 2006 JMLB1 + YUC1-R2 YUC1-F2 + YUC1-R2
yuc2 At4g13260 Salk_030199 Cheng et al, 2006 JMLB1 + YUC2-R1 YUC2-F1 + YUC2-R1
yuc4-1 At5g11320 SM_3_16128 Cheng et al, 2006 SPM32 + YUC4-R2 YUC4-F2 + YUC4-R2
yuc6 At5g25620 Salk_093708 Cheng et al, 2006 YUC6-F3 + JMLB1 YUC6-F3 + YUC6-R3
pdc1-10 At4g33070 Salk_090204 this work JMLB1 + At4g33070-R2 At4g33070-F1 + At4g33070-R2
pdc2-10 At5g54960 Sail_650_C05 this work LB3SAIL + At5g54960-GWR1 At5g54960-F2 + At5g54960-GWR1
pdc3-10 At5g01330 Salk_087974 this work JMLB1 + At5g01330-R2 At5g01330-F2 + At5g01330-R2
pdc4-10 At5g01320 Sail_900_A09 this work LB3SAIL + At5g01320-R2 At5g01320-GWF1 + At5g01320-R2
pdc4-11 At5g01320 Sail_240_G11 this work LB3SAIL + At5g01320-R2 At5g01320-GWF1 + At5g01320-R2
Supplemental Data. Stepanova et al. Plant Cell. (2011). 10.1105/tpc.111.088047
9
Supplemental Table 2. Sequences of the primers used in this study
Primer Name Primer Sequence 24670-F1 5’-GCACGCAAGTGAAGCTCCAAGC-3’ 24670-R1 5’-ATACTGTGGCCAATAGTAAGCC-3’ 70560-F8 5’-CATCAGAGAGACGGTGGTGAAC-3’ 70560-F9 5’-CATGTGCAAAGACATCAGTAACA-3’ 70560-R1 5’-GCTTTTAATGAGCTTCATGTTGG-3’ At4g33070-F1 5'-GCTTCCAAACGGTGACTTGTTATC-3' At4g33070-R2 5'-GCAGGTTCAAATCTTGTGCC-3' At4g33070-GWF1 5’-CACCATGGACACCAAAATCGG-3’ At4g33070-GWR1 5’-CTGAGGATTGGGAGGACG-3’ At5g01320-R2 5'-GCCAAACACAATGTTACTTCAACACC-3' At5g01320-GWF1 5’-CACCATGGACACCAAAATTGGAGCC-3’ At5g01320/30-GWR1 5’-CTGAGGATTGGGAGGACGAC-3’ At5g01330-F2 5'-CCTGTTCTCTTCAAGAATCAAGC-3' At5g01330-R2 5'-CTAGATGTTGACTTTACTAGATGAGC-3' At5g54960-F2 5'-TGCTTCCGATGAAGGTTAGC-3' At5g54960-GWR1 5'-CTGCGGATTTGGGGGACGACTATTAGC-3' At5g54960-GWF1 5’-CACCATGGACACTAAGATCGGATCTATCG-3’ CYP79B2-F 5'-AGTATCATGACCCAATCATCGAC-3' CYP79B2-R 5'-CCATATCGGCTAAGAAGGAC-3' CYP79B3-F 5'-GCAATCCACCAATATCCGTCAG-3' CYP79B3-R 5'-GTTCTATGCATGGACTCGTGG-3' CYP83B1-F 5’-GAGACTCTTGACCCTAACCGC-3’ CYP83B1-R 5’-GCGAGTCCAGTCATGACGTCC-3’ DWLB1 5’-CATACTCATTGCTGATCCATGTAGATTTCC-3’ JMLB1 5’-GGCAATCAGCTGTTGCCCGTCTCACTGGTG-3’ JMLBa 5'-CTTTGACGTTGGAGTCCACGTTC-3' LB3SAIL 5’-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3’ SMP32 5’-TACGAATAAGAGCGTCCATTTTAGAGTGA-3’ TAR1-F1C 5’-CACCATGATGGTTGGGTGTGAAAACTC-3’ TAR1-R1C 5’-AGGCTCGACGCATTTGAGATC-3’ YUC1-F2 5'-TCCGGTTCTCGATGTCGGAG-3' YUC1-R2 5'-GGGTTATGGGAATCATGACGAC-3' YUC2-F1 5'-TTCTTGCATTTTCTCGCTCTA CG-3' YUC2-R1 5'-CTGCATACAATCCGCTTTCGC-3' YUC4-F2 5'-TTGTCAAACCGAGGCGTACC-3' YUC4-R2 5'-ACGACCATATGAGGCAGAGC-3' YUC6-F3 5'-TCTGCAACTTCGGTGCTCAG-3' YUC6-R3 5'-TTTAACGAGAACATCTTAAAGCGCTG-3' WEI8yucc-F1 5'-CACCGGAGGTGGAGGTGGAGCTATGGAGTCTCATCCTCACAACAAAAC-3' WEI8yucc-R1 5'-GGCCCCAGCGGCCGCAGCAGCACCGGATTTAGAGGTAAAGACAAAACG-3' WEI8iaaM-F1 5’-CACCGGAGGTGGAGGTGGAGCTATGTCAGCTTCACCTCTCCTTG-3’ WEI8iaaM-R1 5’-GGCCCCAGCGGCCGCAGCAGCACCATTTCTAGTGCGGTAGTTATATCTC-3’
WEI81stIF1 5’-ACTAACAAATCAGATTTTTATATAGTTTTTCGCTATATTGTTGCAGTGGT GGAGGTGGAGGTGGAGCT-3’
WEI81stIR1 5’-AGTCTTTTATATATAAGGCGTTATATGCGTTATATTCGCTTGCTTATCTA GGCCCCAGCGGCCGCAGCAGCACC-3’