Supplemental Data. Golczyk et al. (2014). Plant Cell …...2014/03/14 · Supplemental Data....
Transcript of Supplemental Data. Golczyk et al. (2014). Plant Cell …...2014/03/14 · Supplemental Data....

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
1
Supplemental Figure 1. Translocation system in Oenothera - Segmental arrangements and inheritance of the Renner complexes. Since the haploid set in Oenothera consists of seven chromosomes, each possessing two ends, chromosomal ends are assigned with numbers from 1 to 14. At the bottom of each figure, each chromosome is represented by a combination of two different digits (separated by a dot). Any two homologous end-segments that conjoin in metaphase I (MI), are designated with the same digit. Segmental arrangement (chromosome formula) in Oenothera is a set of seven pairs of digits, depicting a given Renner complex (seven chromosomes). For example, the bivalent-forming hhookeri de Vries of ancestral Oe. elata ssp. hookeri strain hookeri de Vries was given the reference segmental arrangement 1•2 3•4 5•6 7•8 9•10 11•12 13•14 (a complex*, green, Figure A). The halved colored circles graphically depict genomes composed of the two Renner complexes, each represented by a single circle half. New Renner complexes arise by reciprocal translocation(s) between the non-homologous chromosomes of a haploid set. Although the frequencies of reciprocal translocations are

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
2
difficult to assess, segmental arrangements are stable in culture. Reciprocal translocations occur in micro-evolutionary time frames, probably comparable to spontaneous mutations. For example, chromosomes 1•2 and 3•4 can be involved in a rearrangement leading to a new Renner complex (a’ complex, white) with the chromosomes 2•3 and 4•1 (Figure B). After combining the original Renner complex (a complex) with the new one (a’ complex), a meiotic ring of four (�4, – 1•2 – 2•3 – 3•4 – 4•1– )** plus five bivalents will be formed (Figure B). Extensive crossing experiments and determination of meiotic configurations allowed an assignment of new segmental arrangements in respect to the reference hhookeri de Vries complex, in more than 300 strains. In our example, the two new chromosomes of the Renner complex a’ are given the segmental arrangements 2•3 and 4•1, since they are present in the meiotic ring of four (Figure B). Renner complexes, e.g. a (Figure A), b (Figure C) or c
(Figure D) occur in a homozygotic condition if they lack lethal factors (Figures A,C,D). If segmental arrangements of any two parental complexes are known (Figures A,C,D), one can predict MI configurations in the resulted hybrids (Figures B,E,H,I) and in further generations (Figure F). For example: The a and b complexes (Figures A,C) show extreme translocation heterozygosity – no combination of end-segments is shared between the two complexes: thus, a meiotic ring (�14) arises in the progeny (Figure E). Half of its chromosomes form one linkage group, producing only three possible progeny classes in further generation (Figure F). If the ab (� 14) genotype (Figure E) is a PTH-one, typically aa (7II) and bb (7II) genotypes will be eliminated, and ab will be the only one among the living progeny. The diversity of meiotic configurations is rather a typical situation. Hence new linkage groups can arise upon hybridization and more complicated segregation patterns are expected in further generations (Figure G as an example). This is often the case in natural populations of non-PTH species. In population of PTH species, however, (nearly) complete rings are found. * The above general summary is based on the data reviewed by Cleland (1972), Harte (1994), and Dietrich et al. (1997), cited in the main text. ** For convenience, chains are usually presented instead of rings (Figs. B,E,H,I).

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
3
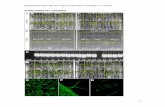
Supplemental Figure 2. A screen for canonical telomeric DNA motifs. (A) – Oe. elata ssp. hookeri strain johansen Standard. Double-target FISH with Arapidopsis-type telomeric probe (left image) and 45S rDNA probe (middle image, arrowheads), right image – DAPI staining. (B) – Asymmetric PCR on known telomeric motifs in Oenothera, Allium, Arabidopsis, and human: 1 kb ladder, Fermentas, Germany (lane 1); lambda/HindIII DNA, Fermentas, Germany (lane 2); Oenothera DNA with Arabidopsis-type, Vertebrate-type, Chlamydomonas-type, Oxytricha-type, Bombyx-type, Tetrahymena-type, and Ascarsis-type telomeric primers (lanes 3-9); Allium DNA as a negative control with the same telomeric primers as employed before (lanes 10-16); and Arabidopsis and human with their own telomeric sequences, as positive controls, respectively (lanes 17, 18). In the positive controls typical high molecular weight smear is observed, whereas primer extension products in all Oenothera
samples were absent (lanes 3-9). Distinct bands that appear for some telomeric motifs in Allium, which lacks telomeric minisatellite repeats, are likely generated by inverted repeats scattered throughout the genome. The lower molecular weight smear may be generated by short interstitial direct repeats (lanes 10-13) (cf. Sykorová et al., 2003; 2006, cited in the main text). Bar = 5 µm.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
4
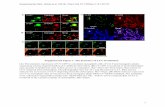
Supplemental Figure 3. Immunodetection of H3S10ph (A,B left images) or H3T11ph (C left image) in root-tip meristem nuclei and chromosomes of Oe. elata ssp. hookeri strain hookeri de Vries. The H3S10ph-foci consistently marked 14 pericentromeres at prophase (A) and later stages (B – anaphase chromosomes with labeled pericentromeres). On the other hand, H3T11ph was distributed along the entire length of condensing mitotic chromosomes (C – metaphase chromosomes), thus confirming the pattern demonstrated by previous authors for other plants. Applying the two labelings allowed to confirm that within the root meristem, mitotic prophases were very rare – about 1 over 600-700 nuclei. Typically, within a microscope view, all the nuclei containing large chromosome-like chromocenters were negative both for H3S10ph and H3T11ph. Arrow in A right points to prophase nucleus. Bars = 5µm.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
5
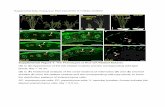
Supplemental Figure 4. Imunonodetection of histone-tail modifications performed on cycling (root-tip meristem) and non-cycling (mature leaf parenchyma) nuclei of Oe. elata ssp. hookeri strain johansen Standard. Immunodetection of H3K9me2 and H3K27me3 gave weak dispersed labeling over the entire nuclear area and no nucleus with a non-random arrangement of fluorescent foci was found (Figures A,B,D,E). Immunodetection of H3K27me2 generated fluorescent foci corresponding to DAPI-positive blocks of terminal heterochromatin (Figure C). Arrowheads indicate DAPI-bright chromatin blocks corresponding to NOR-heterochromatin. Bars = 5µm.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
6
Supplemental Figure 5. Structure of non-cycling nuclei. DAPI-fluorescence (A,B left, D-G) and CMA3-fluorescence (B right, C). (A) – F1 hybrid between Oe. elata ssp. hookeri strain johansen Standard and Oe. grandiflora strain grandiflora Tuscaloosa. Binucleate tapetum cells with nuclei deprived of the facultative chromocenters (left) and individual tapetal nucleus (right, top) with its graphical interpretation (right, bottom), nucleolus – black, NOR-heterochromatin – white. (B) – Oe. biennis strain suaveolens Grado. Mature microspores with nuclei lacking chromocenters (left) or showing the presence of 7-8 small chromocenters (right) much smaller than those present in root meristems. (C) – Oe. elata ssp. hookeri strain johansen Standard. Root hair nuclei without facultative chromocenters, possessing 2 (top image) or 4 (bottom image) smaller chromocenters conforming to NOR-heterochromatin blocks. (D) – Oe.
elata ssp. hookeri strain hookeri de Vries. Hypocotyl parenchyma cells with nuclei deprived of facultative chromocenters, but possessing a few smaller chromocenters conforming to NOR-heterochromatin (arrowheads). Arrow points to the nucleus, which is additionally magnified in the dashed box. (E) – F1 hybrid between Oe. elata ssp. hookeri strain johansen Standard and Oe.
grandiflora strain grandiflora Tuscaloosa. Xylem parenchyma nucleus deprived of large chromocenters, but with two small chromocenters conforming to NOR-heterochromatin (arrows). The adjacent xylem vessel is indicated by an asterisk. (F-G) – Oe. villosa ssp. villosa strain bauri Standard. (F) – a fraction of cotyledon mesophyll cells is characterized by meristematic-type nuclei with distinct large chromocenters (left) while the remaining cells have the differentiated type nuclei – with a clearly visible, but dispersed chromatin meshwork and few smaller chromocenters corresponding in size to the NOR-heterochromatin blocks (right). The young cotyledon mesophyll has still mitotic potential, since mitotically dividing cells were frequently found. Thus, the meristematic-type nuclei are presumably cycing nuclei (G) – Nuclei of root epidermis with chromocenters showing heterogeneity in size and number. The chromocenters are of the size intermediate between this of NOR-heterochromatin blocks and that of large chromocenters from true meristems. It awaits exploration, whether in root epidermis and in a fraction of maturing microspores there is some kind of a selective usage of only some of the DNA sequences, while the rest of them still have to be inert (facultatively condensed). Arrows point to nuclei which are magnified in the dashed boxes. Bars = 5µm.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
7
Supplemental Table 1. Renner complexes (RC), basic genomes (BG), segmental (end) arrangement (SA), and meiotic configurations (MC) in the eight (1-8) Oenothera strains used in this work*.
Species and strain RC BG MC SA** References
1
Oe. elata ssp. hookeri strain
hookeri de Vries
hhookeri de Vries hhookeri de Vries
A
A 7 II
/1·2\ /3·4\ /5·6\ /7·8\ /9·10\ /11·12\ /13·14\
\1·2/ \3·4/ \5·6/ \7·8/ \9·10/ \11·12/ \13·14/ [1-3]
2
Oe. elata ssp. hookeri strain
johansen Standard
hjohansen Standard*** hjohansen Standard
A
A 7 II
/1·2\ /3·4\ /5·6\ /7·10\ /9·8\ /11·12\ /13·14\
\1·2/ \3·4/ \5·6/ \7·10/ \9·8/ \11·12/ \13·14/ [4]
3
Oe. grandiflora strain
grandiflora Tuscaloosa
hgrandiflora Tuscaloosa*** hgrandiflora Tuscaloosa
B
B 7 II
/1·2\ /3·4\ /5·6\ /7·10\ /9·8\ /11·12\ /13·14\
\1·2/ \3·4/ \5·6/ \7·10/ \9·8/ \11·12/ \13·14/ [5]
4
Oe. glazioviana mut.
blandina de Vries****
hblandina de Vries hblandina de Vries
A/B
A/B 7 II
/1·2\ /3·4\ /5·6\ /7·10\ /9·14\ /11·12\ /13·8\
\1·2/ \3·4/ \5·6/ \7·10/ \9·14/ \11·12/ \13·8/ [6; 7; 8]
5
Oe. glazioviana strain
r/r-lamarckiana Sweden*****
r-Svelans
r- Sgaudens
A
B �12, 1 II
/3·4 9·10 13·14 8·5 6·7 11·12 \ /r-1·2\
\ 4·9 10·13 14·8 5·6 7·11 12·3/ \r-1·2/ [2; 3; 8-11]
6
Oe. biennis strain
suaveolens Standard
Stalbicans Stflavens
A
B �12, 1 II
/2·14 13·12 11·10 9·8 7·5 6·3 \ /1·4\
\ 14·13 12·11 10·9 8·7 5·6 3·2/ \1·4/ [2; 3; 8; 12]
7
Oe. biennis strain
suaveolens Grado
Galbicans Gflavens
A
B �14
/1·12 11·10 7·5 6·3 2·14 13·8 9·4 \
\ 12·11 10·7 5·6 3·2 14·13 8·9 4·1/ [13; 14]
8
Oe. villosa ssp. villosa strain
bauri Standard
Stlaxans Stundans
A
A �14
/1·2 3·10 5·8 14·11 12·13 9·6 7·4 \
\ 2·3 10·5 8·14 11·12 13·9 6·7 4·1/ [15; 16]
II – bivalent, � – ring. Segmental arrangements and reciprocal translocations are explained in Supplemental Fig. 1. Renner complexes studied here are of the A or B genome type, or represent a mixture of the A and B genome (A/B). The three “ancestral” homozygotes (1, 2, 3) are putative primitive North American, large-flowered and open-pollinated forms. Four strains (5, 6, 7, 8) represent ring forming species that are permanent translocation heterozygotes.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
8
* Dietrich et al. (1997, cited in the main text) grouped the numerous published PTH and non-PTH species in subsection Oenothera (cf. 17) into 13 distinct species. Five of them are mostly 7-bivalent formers (Oe. elata, Oe. grandiflora, Oe. jamesii, Oe. longissima, Oe. argillicola) and eight ring forming PTH forms (Oe. wolfii, Oe. villosa, Oe. stucchii, Oe. nutans, Oe. biennis, Oe. glazioviana, Oe.
oakesiana, Oe. parviflora). ** Segmental (end) arrangements follow the Cleland’s system (Cleland 1972, cited in the main text). *** The complexes hjohansen Standard and hgrandiflora Tuscaloosa harbor the so called “Johansen segmental arrangement” considered the most ancient in the group. **** Oe. glazioviana mut. blandina de Vries has originated from the famous permanent translocation heterozygote Oe. lamarckiana de Vries (syn. Oe. glazioviana strain lamarckiana de Vries) as a result of crossing experiments and the accompanying spontaneous reciprocal translocations. Its complex hblandina de Vries is a result of translocations between the Renner complexes gaudens and velans with
the consequence of losing the lethal factors (Cleland 1972; Harte 1994, cited in the main text). Hence, this line can be regarded as a “translocation homozygote”. ***** Our Swedish line of Oe. glazioviana is homozygous for recessive allele r present on the bivalent forming chromosome 1·2.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
9
Supplemental Table 2. Average chromosome lengths (CL) and arm ratios (AR) of the measured chromomal types (I – VII) in 7-bivalent forming Oenothera strains.
Oe. elata ssp. hookeri strain hookeri de Vries
Oe. elata ssp. hookeri strain johansen Standard
Oe. grandiflora strain grandiflora Tuscaloosa
Oe. glazioviana strain blandina de Vries
I CL 7.88 (0.34)
AR 1.21 (0.10)
CL 7.81 (0.31) AR 1.12 (0.19)
CL 7.96 (0.32) AR 1.22 (0.19)
CL 7.77 (0.19) AR 1.28 (0.11)
II
CL 7.51 (0.30) AR 1.19 (0.18)
CL 7.62 (0.23) AR 1.22 (0.15)
CL 7.57 (0.24) AR 1.27 (0.17)
CL 7.57 (0.24) AR 1.19 (0.13)
III
CL 7.45 (0.21) AR 1.13 (0.12)
CL 7.35 (0.18) AR 1.09 (0.17)
CL 7.40 (0.22) AR 1.30 (0.19)
CL 7.46 (0.16) AR 1.20 (0.23)
IV
CL 7.26 (0.25) AR 1.17 (0.19)
Cl 7.14 (0.21) AR 1.15 (0.15)
CL 7.00 (0.27) AR 1.19 (0.09)
CL 7.13 (0.33) AR 1.14 (0.21)
V
CL 7.05 (0.27) AR 1.44 (0.18)
CL 6.93 (0.27) AR 1.28 (0.20)
CL 6.92 (0.20) AR 1.32 (0.18)
CL 6.86 (0.19) AR 1.14 (0.18)
VI
CL 7.05 (0.32) AR 1.15 (0.13)
CL 6.90 (0.15) AR 1.15 (0.19)
CL 6.70 (0.40) AR 1.22 (0.12)
CL 6.76 (0.29) AR 1.40 (0.17)
VII
CL 5.80 (0.25) AR 1.22 (0.20)
CL 6.25 (0.29) AR 1.21 (0.18)
CL 6.45 (0.32) AR 1.25 (0.21)
CL 6.45 (0.24) AR 1.13 (0.11)
Standard deviations within brackets. The means are graphically depicted as chromosomal types in Figures 1A- E.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
10
Supplemental Table 3. Mean chromosome length (CL) and arm ratios (AR) of the measured chromosomal types (a - n) in PTH species of Oenothera.
Oe. biennis strain suaveolens Grado
Oe. biennis strain suaveolens Standard
Oe. glazioviana strain r/r-lamarckiana Sweden
Oe. villosa ssp. villosa strain bauri Standard
a CL 8.53 (0.31) AR 1.46 (0.18)
CL 8.68 (0.23) AR 1.57 (0.22)
CL 8.21 (0.18) AR 1.24 (0.09)
CL 7.98 (0.21) AR 1.23 (0.12)
b
CL 8.33 (0.28) AR 1.10 (0.15)
CL 8.38 (0.34) AR 1.15 (0.19)
CL 8.05 (0.23) AR 1.26 (0.11)
CL 7.66 (0.25) AR 1.13 (0.10)
c
CL 8.19 (0.32) AR 1.49 (0.17)
CL 8.00 (0.25) AR 1.41 (0.23)
CL 7.83 (0.26) AR 1.23 ( 0.14)
CL 7.56 (0.23) AR 1.21 (0.16)
d
CL 7.85 (0.20) AR 1.26 (0.09)
CL 7.81 (0.25) AR 1.19 (0.09)
CL 7.45 (0.32) AR 1.34 (0.17)
CL 7.43 (0.22) AR 1.33 (0.15)
e
CL 7.58 (0.23) AR 1.19 (0.11)
CL 7.36 (0.21) AR 1.29 (0.21)
CL 7.43 (0.16) AR 1.36 (0.19)
CL 7.37 (0.19) AR 1.16 (0.14)
f
CL 7.37 (0.20) AR 1.18 (0.09)
CL 7.19 (0.20) AR 1.07 (0.10)
CL 7.37 (0.22) AR 1.27 (0.12)
CL 7.29 (0.24) AR 1.37 (0.11)
g CL 7.14 (0.26) AR 1.46 (0.13)
CL 7.09 (0.24) AR 1.19 (0.15)
CL 7.30 (0.28) AR 2.89 (0.14)
CL 7.26 (0.18) AR 1.25 (0.16)
h
CL 7.08 (0.19) AR 1.30 (0.18)
CL 7.06 (0.16) AR 1.16 (0.13)
CL 7.18 (0.31) AR 1.11 (0.09)
CL 7.23 (0.24) AR 1.33 (0.11)
i
CL 6.82 (0.14) AR 1.32 (0.18)
CL 6.71 (0.15) AR 1.28 (0.21)
CL 6.85 (0.17) AR 1.32 (0.10)
CL 6.98 (0.19) AR 1.34 (0.12)
j
CL 6.78 (0.32) AR 1.34 (0.19)
CL 6.59 (0.18) AR 1.34 (0.18)
CL 6.78 (0.21) AR 1.22 (0.15)
CL 6.89 (0.23) AR 1.08 (0.09)
k
CL 6.37 (0.21) AR 1.21 (0.10)
CL 6.58 (0.13) AR 1.40 (0.07)
CL 6.70 (0.24) AR 1.27 (0.14)
CL 6.71 (0.22) AR 1.42 (0.08)
l
CL 6.11 (0.22) AR 1.28 (0.11)
CL 6.46 (0.14 ) AR 1.24 (0.20)
CL 6.51 (0.26) AR 1.35 (0.10)
CL 6.69 (0.19) AR 1.30 (0.13)
m
CL 6.05 (0.25) AR 1.38 (0.14)
CL 6.41 (0.17) AR 1.34 (0.21)
CL 6.17 (0.22) AR 1.23 (0.11)
CL 6.64 (0.15) AR 1.14 (0.09)
n
CL 5.80 (0.23) AR 1.38 (0.14)
CL 5.68 (0.23) AR 1.47 (0.11)
CL 6.17 (0.28) AR 1.20 (0.13)
CL 6.31 (0.21) AR 1.14 (0.12)
Standard deviations are shown in brackets. The means are graphically depicted as chromosomal types on Figs 1F-I.

Supplemental Data. Golczyk et al. (2014). Plant Cell 10.1105/tpc.114.122655.
11
Supplemental References (references to Supplemental Table 1):
1. de Vries, H (1913). Gruppenweise Artbildung - Unter spezieller Berücksichtigung der Gattung Oenothera. Berlin: Gebrüder Borntraeger;.
2. Cleland, R.E., and Blakeslee, A.F. (1930). Interaction between complexes as evidence for segmental interchange in Oenothera. Proc. Natl. Acad. Sci. 6: 183-189.
3. Cleland, R.E., and Blakeslee, A.F. (1931). Segmental interchange, the basis of chromosomal attachments in Oenothera. Cytologia 2: 175-233.
4. Cleland, R.E. (1935). Cyto-taxonomic studies on certain oenotheras from California. Proc. Amer. Phil. Soc. 75: 339-429.
5. Steiner, E.E., and Stubbe, W. (1984). A contribution to the population biology of Oenothera
grandiflora L’Her. Am. J. Bot. 71:1293-1301. 6. de Vries, H. (1917): Oenothera Lamarckiana mut. velutina. Bot. Gaz. 63: 1-24. 7. de Vries, H. (1923). Über die Entstehung der Oenothera Lamarckiana mut. Veluntina. Biol.
Centralblatt 43: 213-224. 8. Catcheside DG: Structural analysis of Oenothera complexes. Proc Royal Soc Biol Sci Ser B
1940, 128: 509-535. 9. Heribert-Nilsson, N. (1912). Die Variabilität der Oenothera Lamarckiana und das Problem
der Mutation. Z, Ind. Abst. Vererbung. 8:89-231. 10. Emerson, S.H., and Sturtevant, A.H. (1931): Genetical and cytological studies in
Oenothera: III. The translocation interpretation. Z. Ind. Abst. Vererbung 59: 395-419. 11. Renner, O. (1942). Über das Crossing-over bei Oenothera. Flora 136: 117-214. 12. Blaringhem, L. (1914). L’Oenothera Lamackiana Seringe et les Oenothères de la forêt de
Fontainebleau. Rev. Gen. Bot. 25: 35-50. 13. Stubbe, W. (1953) Genetische und zytologische Untersuchungen an verschiedenen Sippen
von Oenothera suaveolens. Z. Ind. Abst. Vererbung 85:180-209. 14. Stubbe, W. and Diers, L. (1958). Die Chromosomenformel des albicans-Komplexes der
Sippe Grado von Oenothera suaveolens. Z. Vererbung 89: 320-322. 15. Renner, O. (1937). Wilde Oenotheren in Norddeutschland. Flora, 31: 182-226. 16. Baerecke, M. (1944). Zur Genetik und Cytologie von Oenothera ammophila Focke, Bauri
Boedijn, Beckeri Renner, parviflora L., rubricaulis Klebahn, silesiaca Renner. Flora 138: 57-92.
17. Rostański, K., Dzhus, M., Gudžinskas, Z., Rostański, A., Shevera, M., and Šulcs, V.V.T. (2004). The Genus Oenothera L. in Eastern Europe. Kraków: W. Szafer Institute of Botany, Polish Academy of Sciences.