Supplemental Data. Lefebvre et al. (2013). Plant Cell 10 ... · Supplemental Data. Lefebvre et al....
Transcript of Supplemental Data. Lefebvre et al. (2013). Plant Cell 10 ... · Supplemental Data. Lefebvre et al....
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Figure 1. Number of introns in four Ustilaginomycotina fungi according to relative position in genes.
0"
100"
200"
300"
400"
500"
600"
700"
800"
900"
1" 2" 3" 4" 5" 6" 7" 8" 9" 10" 11" 12" 13" 14" 15" 16" 17" 18" 19" 20"
Num
ber'of'introns'
Relative'position'in'genes'(5'7>3')'
P.#$locculosa#
U.#maydis#
U.#hordei#
S.#reilianum#
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Figure 2. Relative frequency of introns according to relative position in genes.
0"
0,01"
0,02"
0,03"
0,04"
0,05"
0,06"
0,07"
0,08"
0,09"
0,1"
1" 2" 3" 4" 5" 6" 7" 8" 9" 10" 11" 12" 13" 14" 15" 16" 17" 18" 19" 20"
Relative'fequency'of'introns'
Relative'position'in'genes'(5'7>3')'
P.#$locculosa#
U.#maydis#
U.#hordei#
S.#reilianum#
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Figure 3. Analysis of dinucleotide frequency in P. flocculosa. Dinucleotide frequencies for repeat sequences and gene sequences show no specific depletion of certain pairs; GC-depletion was shown to be a sign of inactivation through repeat induced point mutation (RIP) (Laurie et al., 2012).
-1,2"-1"
-0,8"-0,6"-0,4"-0,2"0"
0,2"0,4"0,6"
AA" AC" AG" AT" CA" CC" CG" CT" GA" GC" GG" GT" TA" TC" TG" TT"
Log 2'(O
bserved/Expected)'
Dinucleotides'
Pf-Repeats"Pf-Genes"
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
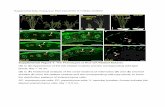
Supplemental Figure 4. Mating assays between P. flocculosa and U. maydis. P. flocculosa and U. maydis strains were tested for their ability to mate. For this assay, haploid cultures were placed as drops on a charcoal-containing medium. Successful mating produces dikaryotic aerial hyphae which appear as a white fluffy colony phenotype. While strong dikaryotic filamentous growth could be observed when U. maydis strains MB215 and FB1 were mated, no aerial hyphae were ever observed when P. flocculosa was mixed with either U. maydis strain or when the strains were selfed. This indicates that P. flocculosa is not able to mate with U. maydis.
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Table 1. Inventory of repetitive sequences in the P. flocculosa genome assembly.
Number of elements*
Length occupied
(bp)
Percentage of
sequence (%)
SINEs 1 68 0.00 ALUs 0 0 0.00 MIRs 0 0 0.00 LINEs 226 22271 0.10 LINE1 13 818 0.00 LINE2 2 174 0.00 L3/CR1 4 421 0.00 LTR elements 413 46452 0.20
ERVL 0 0 0.00
ERVL-MaLRs 0 0 0.00
ERV class I 4 494 0.00
ERV class II 7 359 0.00
DNA elements 266 19267 0.08
hAT-Charlie 0 0 0.00
TcMar-Tigger 0 0 0.00
Unclassified 269 75783 0.32 Total interspersed repeats
163841 0.70
Small RNA 23 5090 0.02 Satellites 18 1583 0.01 Simple repeats 11748 521283 2.23
Low complexity 1913 97309 0.42
* most repeats fragmented by insertions or deletions have been counted as one element
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Table 2. Inventory of repetitive sequences in non-assembled P. flocculosa sequences.
Number of elements*
Length occupied
(bp)
Percentage of
sequence (%)
SINEs 0 0 0.00 ALUs 0 0 0.00 MIRs 0 0 0.00 LINEs 7 782 0.47 LINE1 1 80 0.05 LINE2 0 0 0.00 L3/CR1 0 0 0.00 LTR elements 40 9226 5.55
ERVL
ERVL-MaLRs 0 0 0.00
ERV class I 1 24 0.01
ERV class II 0 0 0.00
DNA elements
hAT-Charlie 0 0 0.00
TcMar-Tigger 0 0 0.00
Unclassified 67 17012 10.24 Total interspersed repeats
27020 16.27
Small RNA 13 3567 2.15 Satellites 0 0 0.00 Simple repeats 121 11098 6.68
Low complexity 13 658 0.40
* most repeats fragmented by insertions or deletions have been counted as one element
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Table 3. OrthoCluster analysis.
Synteny conservation P. flocculosa U. maydis U. hordei U. maydis 0.409 U. hordei 0.484 0.778 S. reilianum 0.308 0.768 0.511
Number of blocks
P. flocculosa U. maydis U. hordei U. maydis 672 U. hordei 683 759 S. reilianum 645 456 612
Largest block size
(Genes*) P. flocculosa U. maydis U. hordei U. maydis 25 U. hordei 34 59 S. reilianum 43 103 87
Average block size
(Genes*) P. flocculosa U. maydis U. hordei U. maydis 4 U. hordei 4 8 S. reilianum 5 14 10
Insertions/deletions
P. flocculosa U. maydis U. hordei U. maydis 12 U. hordei 9 93 S. reilianum 13 125 155
Transpositions
P. flocculosa U. maydis U. hordei U. maydis 67 U. hordei 59 65 S. reilianum 77 38 75
* Only genes from gene families present in both species (homologs) are counted.
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Table 4. Subset of gene correspondence for important biological processes.
Gene Reference gene Gene ID in P. flocculo
sa
BlastP e-value
Mating related genes Pheromone receptor (pra) um02383 pf01711 2.9e-85
Mating pheromone (mfa) S. reilianum16900 pf06826 4.1e-06
Mating pheromone (mfa) UHOR_03899 pf06827 1.7 rba1 um02384 pf01707 2.1e-49 bW um00578 pf03265 5.9e-14 bE um12052 pf03264 3.7e-24 Pheromone response factor (prf) um02713 pf03871 5.8e-51
Ste-20 like protein kinase (smu) um12272 pf06042 0
lga2 (mitochondrial inheritance) AAA99769.1* - -
rga2 (mitochondrial inheritance) AAA99770.1* - -
Transcriptional gene silencing and chromatin remodeling
ARGONAUTE (ago) UHOR_06256 pf06699 0 DICER (dcl) UHOR_08937 pf04802 0 RNA dependent RNA polymerase – (RdRP) UHOR_08874 pf06447 0
RNA dependent RNA polymerase – (RdRP) UHOR_01631 - -
RNA dependent RNA polymerase – (RdRP) UHOR_15740 pf06710 0
Chromodomain-coding HP1-like (Chp) UHOR_05116 - -
Chromodomain-coding HP1-like (Chp) UHOR_07750 pf04343 8.1e-40
Chromodomain-coding (Chp) UHOR_16420 pf06213 1.5e-73
C5-cytosine methyltransferase (DNAme)
UHOR_08509 pf06913 1.1e-100
* Genbank accession number
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental Table 5. Conserved SMURF* secondary metabolite gene clusters.
Type of backbone
gene
ID of backbone gene associated to SMURF cluster
Lowest fraction of shared gene families for all
possible combinations**
Function
1 NRPS pf03176, um10189, UHOR_03116, sr13070
3/5 siderophore biosynthesis (Sid2)
2 NRPS-Like
um01830, UHOR_02719, sr12912
10/16 unknown
3 PKS um10339, sr14030 8/11 unknown
4 PKS um06418, UHOR_08970, sr16861
7/14 unknown
5 NRPS um01434, sr12503 7/13 siderophore biosynthesis (Fer3)
6 PKS pf00007, um06460 5/7 flocculosin/ustilagic acid biosynthesis * Predictions of SMURF (Secondary Metabolite Unique Regions Finder, JCVI) clusters are approximative due to the inclusion of genes not involved in the biosynthesis but physically present in the cluster, or due to the exclusion of genes involved in the biosynthesis but having a function that is not considered by SMURF as a hallmark of secondary metabolite biosynthesis. ** For all combinations of species sharing a SMURF cluster, the lowest proportion of gene families being shared between the clusters of two species is reported.
-
Supplemental Data. Lefebvre et al. (2013). Plant Cell 10.1105/tpc.113.113969
Supplemental References Laurie J.D., Ali S., Linning R., Mannhaupt G., Wong P., Güldener U., Münsterkötter M., Moore R., Kahmann R., Bakkeren G. and Schirawski J. (2012). Genome Comparison of Barley and Maize Smut Fungi Reveals Targeted Loss of RNA Silencing Components and Species-Specific Presence of Transposable Elements. Plant Cell. PMID: 22623492
-
Supplemental*Dataset*1.*Genes*and*gene*families*distribution.
All#genes
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 6238 364 0 0 275U.#maydis 6055 153 350 160 69U.#hordei 5961 166 352 427 205S.#reilianum 5841 239 339 202 52
Fungal#cell#wall#degrading#enzymes
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 39 0 0 0 5U.#maydis 43 0 0 0 1U.#hordei 33 0 0 0 1S.#reilianum 39 0 0 0 0
Plant#cell#wall#degrading#enzymes
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 34 17 0 0 12U.#maydis 28 5 10 6 2U.#hordei 27 8 10 3 0S.#reilianum 27 11 10 4 3
CSEP
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 70 67 0 0 208U.#maydis 77 31 116 89 73U.#hordei 78 25 112 50 103S.#reilianum 81 25 114 125 69
Genes
-
All#genes_fams
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 4950 244 0 0 94U.#maydis 4950 139 289 116 34U.#hordei 4950 81 289 67 53S.#reilianum 4950 193 289 133 19
Fungal#cell#wall#degrading#enzymes_fams
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 22 0 0 0 3U.#maydis 22 0 0 0 1U.#hordei 22 0 0 0 1S.#reilianum 22 0 0 0 0
Plant#cell#wall#degrading#enzymes_fams
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 23 15 0 0 11U.#maydis 23 5 10 5 2U.#hordei 23 8 10 3 0S.#reilianum 23 11 10 4 2
CSEP_fams
Core Shared
Present*exclusively*in*all*three*plant*pathogens
Present*exclusively*in*two*plant*pathogens Species?specific
P.#flocculosa 54 50 0 0 137U.#maydis 54 27 83 70 66U.#hordei 54 22 83 47 75S.#reilianum 54 23 83 93 55
Gene*families
-
Supplemental*Dataset*2.*Inventory*of*genes*related*to*mating*and*pathogenicity.ID Um%Best%Hit%
(*=ortholog)Um%Blastp%evalue
Um%reference
Ustilago%maydis%Database%(MUMDB)%Description Functions%in Gene Proteins%used%in%blastp%to%find%Um%reference
Saccharomyces%cerevisiae%Database%Description%
(SGD)
Meiosis%and%Sporulation%
Specific%Protein
Uh%Best%Hit%(*=ortholog)
Uh%Blastp%evalue
Sr%Best%Hit%(*=ortholog)
Sr%Blastp%evalue
pf05723 um04456* 000E+0 um04456 Adr1*protein*kinase*A,*catalytic*subunit mating*&*pathogenicity*pathways UHOR_06957* 000E+0 sr15343* 000E+0pf02581 um05818* 000E+0 um05818 chimeric*spermidine*synthase/saccharopine*reductase,*required*for*
morphogenesis,*and*indispensable*for*survival*in*the*hostmating*&*pathogenicity*pathways UHOR_08297* 000E+0 sr16440* 000E+0
pf05900 um01514* 000E+0 um01514 dual*specificity*protein*kinase*Fuz7 mating*&*pathogenicity*pathways UHOR_02245* 000E+0 sr12583* 9EU180pf05701 um04474* 000E+0 um04474 Gpa3,*guanine*nucleotideUbinding*protein*alphaU3*subunit mating*&*pathogenicity*pathways UHOR_06981* 000E+0 sr15360* 000E+0pf05889 um01597* 3EU66 um01597 Hap2,*regulates*the*pheromone*response*transcription*factor*prf1 mating*&*pathogenicity*pathways UHOR_02364* 5EU75 sr12660* 20EU75pf03635 um11450* 5EU18 um11450 Hgl1*required*for*dimorphism*and*teliospore*formation mating*&*pathogenicity*pathways UHOR_00981* 60EU33 sr11921* 1EU24pf06405 um03837* 000E+0 um03837 Khd4,*KH*domain*protein*involved*in*cell*growth,*morphogenesis,*
and*pathogenicity,*Rrm4,*RNAUbinding*protein*essential*for*efficient*secretion*of*endochitinase*Cts1
mating*&*pathogenicity*pathways "ORF,*Verified" "YLL032C,*SPBC28E12.02*(SPBC9B6.13)"
"Protein*of*unknown*function*that*may*interact*with*ribosomes,*based*on*coUpurification*experiments;*green*fluorescent*protein*(GFP)Ufusion*protein*localizes*to*the*cytoplasm;*YLL032C*is*not*an*essential*gene"
UHOR_05836* 000E+0 sr14739* 000E+0
pf01651 um02331* 000E+0 um02331 Kpp6*MAP*kinase,*involved*in*fungal*ingress mating*&*pathogenicity*pathways UHOR_03819* 000E+0 sr13532* 000E+0pf02444 um03588* 400EU144 um03588 Med1,*transcription*factor*required*for*mating*and*full*virulence,*
putative*prf1*regulatormating*&*pathogenicity*pathways UHOR_05505* 4EU144 sr14584* 500EU144
pf02701 um05804* 10EU69 um05804 Mzr1,*Cys(2)His(2)*zinc*finger*transcriptional*activator,*regulates*fungal*gene*expression*during*the*biotrophic*growth*stage
mating*&*pathogenicity*pathways UHOR_08284* 200EU66 sr16431* 300EU33
pf04874 um06450* 70EU174 um06450 PkaR/Ubc1*cAMPUdependent*protein*kinase*type*II*regulatory*chain mating*&*pathogenicity*pathways UHOR_09003* 4EU171 sr16888* 5EU180
pf03871 um02713* 300EU54 um02713 Prf1*pheromone*response*factor*Prf1,*Transcription*factor*/*Gene*regulation
mating*&*pathogenicity*pathways STE11*(S.*pombe) SPBC32C12.02 Key*transcription*factor*for*sexual*development
N UHOR_04339* 400EU54 sr13766* 100EU57
pf04304 um10683* 000E+0 um10683 probable*ROK1*U*ATPUdependent*RNA*helicase mating*&*pathogenicity*pathways UHOR_07600* 000E+0 sr15628* 000E+0pf03687 um01643* 3EU96 um01643 Ras2*small*GUprotein mating*&*pathogenicity*pathways UHOR_02437* 2EU96 sr12711* 3EU99pf05613 um03172* 400EU63 um03172 Rbf1,*ZnUfinger*transcription*factor*required*for*bE/bWUactivity mating*&*pathogenicity*pathways UHOR_04948* 3EU60 sr14215* 200EU63pf04653 um11476 3EU27 um11476 related*to*Guanyl*nucleotide*exchange*factor*Sql2 mating*&*pathogenicity*pathways UHOR_07507* 000E+0 sr16103* 000E+0pf03344 um00721* 000E+0 um00721 related*to*MAPKK*kinase mating*&*pathogenicity*pathways UHOR_01105* 000E+0 sr12009* 000E+0pf04966 um03701* 200EU159 um03701 Rok1,*negatively*regulate*morphogenesis*(mating*and*
pathogenicity)*by*controlling*MAPKs*of*the*pheromone*signalling*pathway
mating*&*pathogenicity*pathways UHOR_05655* 300EU156 sr14647* 300EU153
pf02102 um12033* 20EU33 um12033 Rop1*HMGUbox*transcription*factor mating*&*pathogenicity*pathways UHOR_08562* 1EU33 sr12520* 1EU12pf05901 um10803* 000E+0 um10803 Sql2*guanyl*nucleotide*exchange*factor mating*&*pathogenicity*pathways UHOR_02247* 000E+0 sr12585* 000E+0pf00079 um01804* 000E+0 um01804 Tam1,*tryptophan*amino*transferase*(transaminase) mating*&*pathogenicity*pathways UHOR_02680* 000E+0 sr12881* 000E+0pf06573 um03280* 000E+0 um03280 Tup1,*transcriptional*repressor*required*for*dimorphism*and*
virulencemating*&*pathogenicity*pathways UHOR_05109* 000E+0 sr14281* 000E+0
pf03221 um05232* 000E+0 um05232 Uac1*adenylate*cyclase,*Signal*transduction mating*&*pathogenicity*pathways CYR1 YJL005W "Adenylate*cyclase,*required*for*cAMP*production*and*cAMPUdependent*protein*kinase*signaling;*the*cAMP*pathway*controls*a*variety*of*cellular*processes,*including*metabolism,*cell*cycle,*stress*response,*stationary*phase,*and*sporulation"
N UHOR_03218* 000E+0 sr13141* 000E+0
pf05327 um05261* 000E+0 um05261 Ubc2*MAP*kinase*pathwayUinteracting*protein mating*&*pathogenicity*pathways UHOR_08070* 000E+0 sr16309* 000E+0pf05549 um03305* 000E+0 um03305 Ubc3/Kpp2*MAP*kinase mating*&*pathogenicity*pathways UHOR_05144* 000E+0 sr14305* 000E+0pf06027 um04258.2* 000E+0 um04258.2 Ubc4/Kpp4*MAPKK*kinase mating*&*pathogenicity*pathways UHOR_06391* 000E+0 sr15150* 000E+0pf02900 um10375 2EU18 um10375 UmRrm75,*related*to*HRB1*U*Poly(A+)*RNAUbinding*protein,*involved*
in*the*export*of*mRNAs*from*the*nucleus*to*the*cytoplasmmating*&*pathogenicity*pathways UHOR_04825 600EU21 sr14141 40EU21
pf05965 um03924 40EU12 um03924 Rep1,*repellent*protein*1*precursor pathogenicity? N UHOR_05948 1EU12 sr14829 2EU12pf02509 um05543 40EU81 um05543 Don3,*Ste20Ulike*kinase*triggers*septin*reorganization*during*
secondary*septum*formationpathogenicity? UHOR_03730* 000E+0 sr13462* 000E+0
-
Supplemental*Dataset*3.*Inventory*of*genes*related*to*meiosis*and*other*functions.Legend
Meiosis&and&Sporulation&Other&functionsID Um&Best&Hit&(*=ortholog)
Um&Blastp&evalue
Um&reference
Ustilago&maydis&Database&(MUMDB)&Description Functions&in Gene Proteins&used&in&blastp&to&find&Um&reference
Saccharomyces&cerevisiae&Database&Description&(SGD) Meiosis&and&Sporulation&
Specific&Protein
Uh&Best&Hit&(*=ortholog)
Uh&Blastp&evalue Sr&Best&Hit&(*=ortholog)
Sr&Blastp&evalue
pf00378 um00651* 000E+0 um00651 related*to*CDC23*F*cell*division*control*protein AnaphaseFpromoting*complex CDC23 "YHR166C,*SPAC6F12.14" "Subunit*of*the*anaphaseFpromoting*complex/cyclosome*(APC/C),*which*is*a*ubiquitinFprotein*ligase*required*for*degradation*of*anaphase*inhibitors,*including*mitotic*cyclins,*during*the*metaphase/anaphase*transition"
N UHOR_01001* 000E+0 sr11932* 000E+0
pf03682 um01647* 000E+0 um01647 related*to*CDC20*F*cell*division*control*protein AnaphaseFpromoting*complex CDC20 "YGL116W,*SPAC821.08c" "CellFcycle*regulated*activator*of*anaphaseFpromoting*complex/cyclosome*(APC/C),*which*is*required*for*metaphase/anaphase*transition;*directs*ubiquitination*of*mitotic*cyclins,*Pds1p,*and*other*anaphase*inhibitors;*potential*Cdc28p*substrate"
N UHOR_02442* 000E+0 sr12715* 000E+0
pf00610 um02089* 000E+0 um02089 related*to*nuclear*protein*bimA AnaphaseFpromoting*complex CDC27 "YBL084C,*SPAC17C9.01c*(SPAC1851.01)" "Subunit*of*the*AnaphaseFPromoting*Complex/Cyclosome*(APC/C),*which*is*a*ubiquitinFprotein*ligase*required*for*degradation*of*anaphase*inhibitors,*including*mitotic*cyclins,*during*the*metaphase/anaphase*transition"
N UHOR_03468* 10EF135 sr13311* 100EF135
pf01760 um02427* 000E+0 um02427 related*to*negative*regulator*of*mitosis AnaphaseFpromoting*complex APC1 "YNL172W,*SPBC106.09" "Largest*subunit*of*the*AnaphaseFPromoting*Complex/Cyclosome*(APC/C),*which*is*a*ubiquitinFprotein*ligase*required*for*degradation*of*anaphase*inhibitors,*including*mitotic*cyclins,*during*the*metaphase/anaphase*transition"
N UHOR_03971* 000E+0 sr10844.2* 000E+0
pf00824 um04059* 000E+0 um04059 related*to*anaphase*control*protein*cut9 AnaphaseFpromoting*complex CDC16 "YKL022C,*SPAC6F12.15c,*NCU01377.2" "Subunit*of*the*anaphaseFpromoting*complex/cyclosome*(APC/C),*which*is*a*ubiquitinFprotein*ligase*required*for*degradation*of*anaphase*inhibitors,*including*mitotic*cyclins,*during*the*metaphase/anaphase*transition;*required*for*sporulation"
N UHOR_06138* 000E+0 sr14968* 000E+0
pf00131 um10036* 4EF135 um10036 conserved*hypothetical*protein AnaphaseFpromoting*complex APC5 "YOR249C,*SPAC959.09c" "Subunit*of*the*AnaphaseFPromoting*Complex/Cyclosome*(APC/C),*which*is*a*ubiquitinFprotein*ligase*required*for*degradation*of*anaphase*inhibitors,*including*mitotic*cyclins,*during*the*metaphase/anaphase*transition"
N UHOR_00231* 7EF138 sr11491* 300EF141
pf05866 um10809.2* 600EF12 um10809.2 hypothetical*protein AnaphaseFpromoting*complex CDC26 "YFR036W,*SPAC23C11.12*" "Subunit*of*the*AnaphaseFPromoting*Complex/Cyclosome*(APC/C),*which*is*a*ubiquitinFprotein*ligase*required*for*degradation*of*anaphase*inhibitors,*including*mitotic*cyclins,*during*the*metaphase/anaphase*transition"
N UHOR_02282* 100EF9 sr12610* 30EF12
pf03354 um11615.2* 50EF63 um11615.2 related*to*anaphase*promoting*complex*subunit*10 AnaphaseFpromoting*complex DOC1 "YGL240W,*NCU08731.2" "Processivity*factor*required*for*the*ubiquitination*activity*of*the*anaphase*promoting*complex*(APC),*mediates*the*activity*of*the*APC*by*contributing*to*substrate*recognition;*involved*in*cyclin*proteolysis"
N UHOR_01047* 2EF54 sr11967* 10EF57
pf05533 um03234.2* 000E+0 um03234.2 related*to*CDC5*F*Serine/threonineFprotein*kinase Cell*cycle*regulators CDC5 "YMR001C,*SPAC23C11.16" "PoloFlike*kinase*with*similarity*to*Xenopus*Plx1*and*S.*pombe*Plo1p;*found*at*bud*neck,*nucleus*and*SPBs;*has*multiple*functions*in*mitosis*and*cytokinesis*through*phosphorylation*of*substrates;*may*be*a*Cdc28p*substrate"
N UHOR_05036* 000E+0 sr14229* 000E+0
pf01181 um03758* 2EF159 um03758 Clb1*bFtype*cyclin*1 Cell*cycle*regulators CLB5 "YPR120C,*NCU02758.1" BFtype*cyclin*involved*in*DNA*replication*during*S*phase;*activates*Cdc28p*to*promote*initiation*of*DNA*synthesis;*functions*in*formation*of*mitotic*spindles*along*with*Clb3p*and*Clb4p;*most*abundant*during*late*G1*phase
N UHOR_05723* 3EF159 sr11244* 10EF168
pf05129 um05574* 10EF72 um05574 conserved*hypothetical*protein Cell*cycle*regulators CLG1 "YGL215W,*SPBC1D7.03" CyclinFlike*protein*that*interacts*with*Pho85p;*has*sequence*similarity*to*G1*cyclins*PCL1*and*PCL2 N UHOR_07990* 30EF102 sr16259* 600EF93pf06796 um06187* 40EF159 um06187 related*to*CDC14*F*dual*specificity*phosphatase Cell*cycle*regulators CDC14 "YFR028C,*SPAC1782.09c" "Protein*phosphatase*required*for*mitotic*exit;*located*in*the*nucleolus*until*liberated*by*the*FEAR*and*
Mitotic*Exit*Network*in*anaphase,*enabling*it*to*act*on*key*substrates*to*effect*a*decrease*in*CDK/BFcyclin*activity*and*mitotic*exit"
N UHOR_08766* 100EF159 sr11301* 300EF162
pf00397 um00172* 20EF78 um00172 related*to*meiotic*recombination*protein*rec8 Chromosome*cohesion REC8 "YPR007C,*SPBC29A10.14" MeiosisFspecific*component*of*sister*chromatid*cohesion*complex;*maintains*cohesion*between*sister*chromatids*during*meiosis*I;*maintains*cohesion*between*centromeres*of*sister*chromatids*until*meiosis*II;*homolog*of*S.*pombe*Rec8p
Y UHOR_00276* 3EF84 sr11519* 100EF87
pf05709 um04389* 000E+0 um04389 smc3*probable*SMC3*F*required*for*structural*maintenance*of*chromosomes
Chromosome*cohesion SMC3 "YJL074C,*SPAC10F6.09c" "Subunit*of*the*multiprotein*cohesin*complex*required*for*sister*chromatid*cohesion*in*mitotic*cells;*also*required,*with*Rec8p,*for*cohesion*and*recombination*during*meiosis;*phylogenetically*conserved*SMC*chromosomal*ATPase*family*member"
N UHOR_06857* 000E+0 sr15276* 000E+0
pf04551 um04677* 2EF45 um04677 putative*protein Chromosome*segregation SGO1 YOR073W "Component*of*the*spindle*checkpoint,*involved*in*sensing*lack*of*tension*on*mitotic*chromosomes;*protects*centromeric*Rec8p*at*meiosis*I;*required*for*accurate*chromosomal*segregation*at*meiosis*II*and*for*mitotic*chromosome*stability"
N UHOR_06729* 800EF78 sr15563* 500EF51
pf03004 um10782* 000E+0 um10782 related*to*STU2*F*MicrotubuleFassociated*protein*(MAP)*of*the*XMAP215/Dis1*family
Chromosome*segregation STU2 "YLR045C,*SPCC736.14" MicrotubuleFassociated*protein*(MAP)*of*the*XMAP215/Dis1*family;*regulates*microtubule*dynamics*during*spindle*orientation*and*metaphase*chromosome*alignment;*interacts*with*spindle*pole*body*component*Spc72p
N UHOR_03349* 000E+0 sr13224* 000E+0
pf00631 um10958* 000E+0 um10958 related*to*kinetochore*associated*2*(HEC) Chromosome*segregation TID3 "YIL144W,*SPBC11C11.03" "Component*of*the*evolutionarily*conserved*kinetochoreFassociated*Ndc80*complex*(Ndc80pFNuf2pFSpc24pFSpc25p);*conserved*coiledFcoil*protein*involved*in*chromosome*segregation,*spindle*checkpoint*activity,*kinetochore*assembly*and*clustering"
N UHOR_00391* 000E+0 sr11601* 000E+0
pf02331 um01110* 000E+0 um01110 related*to*serineFprotein*kinase*atr DNA*damage*checkpoint MEC1 YBR136W Genome*integrity*checkpoint*protein*and*PI*kinase*superfamily*member;*signal*transducer*required*for*cell*cycle*arrest*and*transcriptional*responses*prompted*by*damaged*or*unreplicated*DNA;*monitors*and*participates*in*meiotic*recombination
N UHOR_01678* 000E+0 sr12409* 000E+0
pf03738 um01605* 1EF138 um01605 related*to*cell*cycle*checkpoint*protein*RAD17 DNA*damage*checkpoint RAD24 YER173W "Checkpoint*protein,*involved*in*the*activation*of*the*DNA*damage*and*meiotic*pachytene*checkpoints;*subunit*of*a*clamp*loader*that*loads*Rad17pFMec3pFDdc1p*onto*DNA;*homolog*of*human*and*S.*pombe*Rad17*protein"
N UHOR_02377* 200EF144 sr12671* 300EF144
pf06539 um11016* 3EF78 um11016 Rec1*exonuclease*REC1 DNA*damage*checkpoint RAD17 YOR368W "Checkpoint*protein,*involved*in*the*activation*of*the*DNA*damage*and*meiotic*pachytene*checkpoints;*with*Mec3p*and*Ddc1p,*forms*a*clamp*that*is*loaded*onto*partial*duplex*DNA;*homolog*of*human*and*S.*pombe*Rad1*and*U.*maydis*Rec1*proteins"
N UHOR_05243* 50EF78 sr14379* 1EF78
pf03356 um00584* 000E+0 um00584 probable*casein*kinaseF1*hhp1 DNA*Repair HRR25 "YPL204W,*SPBC3H7.15" "Protein*kinase*involved*in*regulating*diverse*events*including*vesicular*trafficking,*DNA*repair,*and*chromosome*segregation;*binds*the*CTD*of*RNA*pol*II;*homolog*of*mammalian*casein*kinase*1delta*(CK1delta)"
N UHOR_00903* 000E+0 sr11862* 000E+0
pf05517 um03141* 20EF171 um03141 related*to*EXO1*F*exonuclease*which*interacts*with*Msh2p
DNA*Repair EXO1 "YOR033C,*SPBC29A10.05" "5'F3'*exonuclease*and*flapFendonuclease*involved*in*recombination,*doubleFstrand*break*repair*and*DNA*mismatch*repair;*member*of*the*Rad2p*nuclease*family,*with*conserved*N*and*I*nuclease*domains"
N UHOR_04910* 200EF153 sr11071* 7EF171
pf03154 um05228* 50EF57 um05228 related*to*RAD23*F*nucleotide*excision*repair*protein*(ubiquitinFlike*protein)
DNA*Repair RAD23 "YEL037C,*SPBC2D10.12" "Protein*with*ubiquitinFlike*N*terminus,*recognizes*and*binds*damaged*DNA*(with*Rad4p)*during*nucleotide*excision*repair;*regulates*Rad4p*levels,*subunit*of*Nuclear*Excision*Repair*Factor*2*(NEF2);*homolog*of*human*HR23A*and*HR23B*proteins"
N UHOR_03213* 3EF51 sr13137* 700EF96
pf04168 um01271* 100EF132 um01271 related*to*NAM8*F*meiotic*recombination*protein DSB*generation NAM8 "YHR086W,*NCU00768.1" "RNA*binding*protein,*component*of*the*U1*snRNP*protein;*mutants*are*defective*in*meiotic*recombination*and*in*formation*of*viable*spores,*involved*in*the*formation*of*DSBs*through*meiosisFspecific*splicing*of*MER2*preFmRNA"
N UHOR_01909* 30EF126 sr10390* 4EF123
pf06020 um10420 300EF9 um10420 related*to*meiotic*recombination*protein*SPO11 DSB*generation SPO11 "YHL022C,*SPAC17A5.11,*NCU01120.2" MeiosisFspecific*protein*that*initiates*meiotic*recombination*by*catalyzing*the*formation*of*doubleFstrand*breaks*in*DNA*via*a*transesterification*reaction;*required*for*homologous*chromosome*pairing*and*synaptonemal*complex*formation
Y UHOR_06398 20EF9 sr15155* 7EF12
pf06021 um10420* 70EF9 um10420 related*to*meiotic*recombination*protein*SPO11 DSB*generation SPO11 "YHL022C,*SPAC17A5.11,*NCU01120.2" MeiosisFspecific*protein*that*initiates*meiotic*recombination*by*catalyzing*the*formation*of*doubleFstrand*breaks*in*DNA*via*a*transesterification*reaction;*required*for*homologous*chromosome*pairing*and*synaptonemal*complex*formation
Y UHOR_06398* 8EF9 sr15155 10EF9
pf06526 um11008* 000E+0 um11008 related*to*HFM1*F*DNA/RNA*helicase DSB*generation MER3 "YGL251C,*NCU09793.2" Meiosis*specific*DNA*helicase*involved*in*the*conversion*of*doubleFstranded*breaks*to*later*recombination*intermediates*and*in*crossover*control;*catalyzes*the*unwinding*of*Holliday*junctions;*has*ssDNA*and*dsDNA*stimulated*ATPase*activity
N UHOR_05215* 000E+0 sr14355* 000E+0
pf03249 um11201.2* 50EF96 um11201.2 putative*protein DSB*generation REC107 YJR021C Protein*involved*in*early*stages*of*meiotic*recombination;*involved*in*coordination*between*the*initiation*of*recombination*and*the*first*division*of*meiosis;*part*of*a*complex*(Rec107pFMei4pFRec114p)*required*for*ds*break*formation
Y UHOR_01171* 30EF96 sr12057* 100EF108
pf03761 um01932* 2EF165 um01932 related*to*PMS1*F*DNA*mismatch*repair*protein Mismatch*repair PMS1 "YNL082W,*NCU08020.1" "ATPFbinding*protein*required*for*mismatch*repair*in*mitosis*and*meiosis;*functions*as*a*heterodimer*with*Mlh1p,*binds*doubleF*and*singleFstranded*DNA*via*its*NFterminal*domain,*similar*to*E.*coli*MutL"
N UHOR_02869* 000E+0 sr10676* 000E+0
pf01208 um03025* 000E+0 um03025 probable*DNA*mismatch*repair*protein*MSH2 Mismatch*repair MSH2 "YOL090W,*NCU02230.2" Protein*that*forms*heterodimers*with*Msh3p*and*Msh6p*that*bind*to*DNA*mismatches*to*initiate*the*mismatch*repair*process;*contains*a*Walker*ATPFbinding*motif*required*for*repair*activity;*Msh2pFMsh6p*binds*to*and*hydrolyzes*ATP
N UHOR_04750* 000E+0 sr14085* 000E+0
pf05625 um11009* 000E+0 um11009 related*to*MSH6*F*DNA*mismatch*repair*protein Mismatch*repair MSH6 "YDR097C,*NCU08135.1" "Protein*required*for*mismatch*repair*in*mitosis*and*meiosis,*forms*a*complex*with*Msh2p*to*repair*both*singleFbase*&*insertionFdeletion*mispairs;*potentially*phosphorylated*by*Cdc28p"
N UHOR_05217* 000E+0 sr14354* 000E+0
pf03074 um05148* 000E+0 um05148 "related*to*ATPFdependent*DNA*helicase*II,*70*kDa*subunit"
Nonhomologous*end*joining YKU70 "YMR284W,*NCU08290.1" "Subunit*of*the*telomeric*Ku*complex*(Yku70pFYku80p),*involved*in*telomere*length*maintenance,*structure*and*telomere*position*effect;*relocates*to*sites*of*doubleFstrand*cleavage*to*promote*nonhomologous*end*joining*during*DSB*repair"
N UHOR_03093* 000E+0 sr13053* 000E+0
pf00286 um00099* 1EF162 um00099 related*to*cholineFphosphate*cytidylyltransferase other PCT1 "YGR202C,*SPCC1827.02c" "Cholinephosphate*cytidylyltransferase,*also*known*as*CTP:phosphocholine*cytidylyltransferase,*rateFdetermining*enzyme*of*the*CDPFcholine*pathway*for*phosphatidylcholine*synthesis,*inhibited*by*Sec14p,*activated*upon*lipidFbinding"
N UHOR_00160* 2EF177 sr11438* 200EF159
pf06224 um00469* 3EF60 um00469 probable*histone*H2A*F/Z*family*member*HTZ1 other HTZ1 "YOL012C,*SPBC11B10.10c" "Histone*variant*H2AZ,*exchanged*for*histone*H2A*in*nucleosomes*by*the*SWR1*complex;*involved*in*transcriptional*regulation*through*prevention*of*the*spread*of*silent*heterochromatin"
N UHOR_00733* 70EF63 sr10150* 40EF63
pf06217 um00486* 000E+0 um00486 pgm2*probable*PGM2*F*phosphoglucomutase other PGM2 "YMR105C,*SPBC32F12.10" "Phosphoglucomutase,*catalyzes*the*conversion*from*glucoseF1Fphosphate*to*glucoseF6Fphosphate,*which*is*a*key*step*in*hexose*metabolism;*functions*as*the*acceptor*for*a*GlcFphosphotransferase"
N UHOR_00757* 000E+0 sr10174* 000E+0
-
pf03486 um00756* 000E+0 um00756 gdi1*probable*GDI1*F*GDP*dissociation*inhibitor other GDI1 "YER136W,*SPAC22H10.12c" "GDP*dissociation*inhibitor,*regulates*vesicle*traffic*in*secretory*pathways*by*regulating*the*dissociation*
of*GDP*from*the*Sec4/Ypt/rab*family*of*GTP*binding*proteins"
N UHOR_01157* 000E+0 sr12047* 000E+0
pf02813 um00851* 000E+0 um00851 related*to*BFR1*F*Nuclear*segregation*protein other BFR1 YOR198C Component*of*mRNP*complexes*associated*with*polyribosomes;*implicated*in*secretion*and*nuclear*
segregation;*multicopy*suppressor*of*BFA*(Brefeldin*A)*sensitivity
N UHOR_01279* 2EF177 sr12141* 000E+0
pf01108 um00983* 000E+0 um00983 conserved*hypothetical*protein other SNT1 YCR033W "Subunit*of*the*Set3C*deacetylase*complex*that*interacts*directly*with*the*Set3C*subunit,*Sif2p;*putative*
DNAFbinding*protein"
N UHOR_01495* 000E+0 sr12279* 400EF12
pf02961 um01221* 000E+0 um01221 Tub1*alpha*tubulin other TUB3 YML124C "AlphaFtubulin;*associates*with*betaFtubulin*(Tub2p)*to*form*tubulin*dimer,*which*polymerizes*to*form*
microtubules;*expressed*at*lower*level*than*Tub1p"
N UHOR_01843* 000E+0 sr10296* 000E+0
pf05805 um01561* 000E+0 um01561 probable*Pelota*protein other DOM34 YNL001W "Endoribonuclease;*functions*in*noFgo*mRNA*decay,*protein*translation*to*promote*G1*progression*and*
differentiation,*required*for*meiotic*cell*division;*similar*to*the*eukaryotic*Pelota"
N UHOR_02320* 000E+0 sr12633* 000E+0
pf00423 um02016* 1EF78 um02016 related*to*Membrane*steroid*binding*protein other DAP1 "YPL170W,*SPAC25B8.01*(SPAC26H5.15)" "HemeFbinding*protein*involved*in*regulation*of*cytochrome*P450*protein*Erg11p;*damage*response*
protein,*related*to*mammalian*membrane*progesterone*receptors;*mutations*lead*to*defects*in*
telomeres,*mitochondria,*and*sterol*synthesis"
N UHOR_02994* 6EF69 sr12977* 20EF78
pf03670 um02065* 000E+0 um02065 Hda1*histone*deacetylase other RPD3 YNL330C Histone*deacetylase;*regulates*transcription*and*silencing N UHOR_03433* 000E+0 sr13286* 000E+0
pf02023 um02607* 000E+0 um02607 probable*ISW2*F*ATPase*component*of*a*two*subunit*
chromatin*remodeling*complex
other ISW2 YOR304W Member*of*the*imitationFswitch*(ISWI)*class*of*ATPFdependent*chromatin*remodeling*complexes N UHOR_04165* 000E+0 sr13646* 000E+0
pf03975 um02703* 000E+0 um02703 "fbp1*probable*FBP1*F*fructoseF1,6Fbisphosphatase" other FBP1 "YLR377C,*SPBC1198.14c" "FructoseF1,6Fbisphosphatase,*key*regulatory*enzyme*in*the*gluconeogenesis*pathway,*required*for*
glucose*metabolism"
N UHOR_04313* 000E+0 sr13755* 000E+0
pf02234 um03470* 000E+0 um03470 related*to*putative*calcium*PFtype*ATPase*NCAF2 other PMC1 "YGL006W,*SPAPB2B4.04c" Vacuolar*Ca2+*ATPase*involved*in*depleting*cytosol*of*Ca2+*ions;*prevents*growth*inhibition*by*
activation*of*calcineurin*in*the*presence*of*elevated*concentrations*of*calcium;*similar*to*mammalian*
PMCA1a
N UHOR_05344* 000E+0 sr14461* 000E+0
pf01250 um03717* 000E+0 um03717 conserved*hypothetical*protein other GSG1 YDR108W "Subunit*of*TRAPP*(transport*protein*particle),*a*multiFsubunit*complex*involved*in*targeting*and/or*
fusion*of*ERFtoFGolgi*transport*vesicles*with*their*acceptor*compartment;*protein*has*late*meiotic*role,*
following*DNA*replication"
N UHOR_05675* 000E+0 sr14655* 000E+0
pf00738 um03958* 4EF129 um03958 fen1*probable*FEN1*F*fatty*acid*elongase other SUR4 "YLR372W,*SPAC1B2.03c" "Elongase,*involved*in*fatty*acid*and*sphingolipid*biosynthesis;*synthesizes*very*long*chain*20F26Fcarbon*
fatty*acids*from*C18FCoA*primers;*involved*in*regulation*of*sphingolipid*biosynthesis"
N UHOR_05999* 500EF132 sr14863* 2EF135
pf00963 um03994* 000E+0 um03994 "pdc1*probable*PDC1*F*pyruvate*decarboxylase,*
isozyme*1"
other PDC1 "YLR044C,*SPAC13A11.06" "Major*of*three*pyruvate*decarboxylase*isozymes,*key*enzyme*in*alcoholic*fermentation,*decarboxylates*
pyruvate*to*acetaldehyde;*subject*to*glucoseF,*ethanolF,*and*autoregulation;*involved*in*amino*acid*
catabolism"
N UHOR_06047* 000E+0 sr14897* 000E+0
pf00769 um04064* 1EF171 um04064 related*to*Glucoamylase*precursor other SGA1 "YIL099W,*SPBC14C8.05c" "Intracellular*sporulationFspecific*glucoamylase*involved*in*glycogen*degradation;*induced*during*
starvation*of*a/a*diploids*late*in*sporulation,*but*dispensable*for*sporulation"
N UHOR_06142* 000E+0 sr14974* 000E+0
pf00882 um04081* 4EF90 um04081 related*to*glutathione*SFtransferase other GTT1 "YIR038C,*SPAC688.04c" "ER*associated*glutathione*SFtransferase*capable*of*homodimerization;*expression*induced*during*the*
diauxic*shift*and*throughout*stationary*phase;*functional*overlap*with*Gtt2p,*Grx1p,*and*Grx2p"
N UHOR_06159* 600EF96 sr14992* 400EF99
pf06735 um04159* 400EF126 um04159 rki1*probable*RKI1*F*DFriboseF5Fphosphate*ketolF
isomerase
other RKI1 "YOR095C,*SPAC144.12" "RiboseF5Fphosphate*ketolFisomerase,*catalyzes*the*interconversion*of*ribose*5Fphosphate*and*ribulose*
5Fphosphate*in*the*pentose*phosphate*pathway;*participates*in*pyridoxine*biosynthesis"
N UHOR_06220* 70EF123 sr15039* 500EF120
pf06077 um04228* 600EF156 um04228 related*to*SSO1*F*syntaxinFrelated*protein other SSO2 "YMR183C,*SPCC825.03c" Plasma*membrane*tFSNARE*involved*in*fusion*of*secretory*vesicles*at*the*plasma*membrane;*syntaxin*
homolog*that*is*functionally*redundant*with*Sso1p
N UHOR_06348* 2EF144 sr15115* 5EF147
pf01396 um04590* 50EF132 um04590 related*to*CHO1*F*CDPFdiacylglycerol*serine*OF
phosphatidyltransferase
other CHO1 "YER026C,*SPCC1442.12" "Phosphatidylserine*synthase,*functions*in*phospholipid*biosynthesis;*catalyzes*the*reaction*CDPF
diaclyglycerol*+*LFserine*=*CMP*+*LF1Fphosphatidylserine,*transcriptionally*repressed*by*myoFinositol*and*
choline"
N UHOR_06604* 2EF102 sr15473* 100EF135
pf02925 um04960* 500EF66 um04960 related*to*30S*ribosomal*protein*S12 other "ORF,*
Verified"
"YNR036C,*SPAC4F8.06" Mitochondrial*protein;*may*interact*with*ribosomes*based*on*coFpurification*experiments;*similar*to*E.*
coli*and*human*mitochondrial*S12*ribosomal*proteins
N UHOR_07096* 100EF66 sr15840* 1EF66
pf05361 um05355* 500EF141 um05355 related*to*acylFCoA*sterol*acyltransferase other ARE2 "YNR019W,*SPAC13G7.05" "AcylFCoA:sterol*acyltransferase,*isozyme*of*Are1p;*endoplasmic*reticulum*enzyme*that*contributes*the*
major*sterol*esterification*activity*in*the*presence*of*oxygen"
N UHOR_08178* 7EF138 sr16338* 000E+0
pf05134 um05567* 4EF63 um05567 atg8*probable*ATG8*F*essential*for*autophagy other AUT7 "YBL078C,*SPBP8B7.24c" "Protein*required*for*autophagy;*modified*by*the*serial*action*of*Atg4p,*Atg7p,*and*Atg3p,*and*
conjugated*at*the*C*terminus*with*phosphatidylethanolamine,*to*become*the*form*essential*for*
generation*of*autophagosomes"
N UHOR_07982* 5EF63 sr16253* 4EF63
pf02646 um05828* 000E+0 um05828 probable*tubulin*beta*chain other TUB2 YFL037W "BetaFtubulin;*associates*with*alphaFtubulin*(Tub1p*and*Tub3p)*to*form*tubulin*dimer,*which*
polymerizes*to*form*microtubules"
N UHOR_08314* 000E+0 sr16450* 000E+0
pf00465 um10050* 2EF42 um10050 probable*U6*snRNAFassociated*SmFlike*protein*LSm5 other LSM5 YER146W "Lsm*(Like*Sm)*protein;*part*of*heteroheptameric*complexes*(Lsm2pF7p*and*either*Lsm1p*or*8p):*
cytoplasmic*Lsm1p*complex*involved*in*mRNA*decay;*nuclear*Lsm8p*complex*part*of*U6*snRNP*and*
possibly*involved*in*processing*tRNA,*snoRNA,*and*rRNA"
N UHOR_00303* 300EF42 sr11537* 20EF42
pf03300 um10117*,um
04290*,um101
20*
000E+0 um10117 Chs4*chitin*Synthase*4 other CHS1 "YNL192W,*SPAC13G6.12c" "Chitin*synthase*I,*requires*activation*from*zymogenic*form*in*order*to*catalyze*the*transfer*of*NF
acetylglucosamine*(GlcNAc)*to*chitin;*required*for*repairing*the*chitin*septum*during*cytokinesis;*
transcription*activated*by*mating*factor"
N UHOR_00740*,U
HOR_06435*,UH
OR_00723*
000E+0 sr15181*,sr1
0158*,sr1013
9*
000E+0
pf04843 um10117*,um
10718.2*
000E+0 um10117 Chs4*chitin*Synthase*4 other CHS1 "YNL192W,*SPAC13G6.12c" "Chitin*synthase*I,*requires*activation*from*zymogenic*form*in*order*to*catalyze*the*transfer*of*NF
acetylglucosamine*(GlcNAc)*to*chitin;*required*for*repairing*the*chitin*septum*during*cytokinesis;*
transcription*activated*by*mating*factor"
N UHOR_07282* 000E+0 sr16010* 000E+0
pf06153 um10117*,um
04290*,um107
18.2*,um1012
0*
000E+0 um10117 Chs4*chitin*Synthase*4 other CHS1 "YNL192W,*SPAC13G6.12c" "Chitin*synthase*I,*requires*activation*from*zymogenic*form*in*order*to*catalyze*the*transfer*of*NF
acetylglucosamine*(GlcNAc)*to*chitin;*required*for*repairing*the*chitin*septum*during*cytokinesis;*
transcription*activated*by*mating*factor"
N UHOR_00740*,U
HOR_00723*,UH
OR_06435*
000E+0 sr15181*,sr1
6010,sr10158
*,sr10139*
000E+0
pf06222 um10117*,um
04290*,um101
20*
000E+0 um10117 Chs4*chitin*Synthase*4 other CHS1 "YNL192W,*SPAC13G6.12c" "Chitin*synthase*I,*requires*activation*from*zymogenic*form*in*order*to*catalyze*the*transfer*of*NF
acetylglucosamine*(GlcNAc)*to*chitin;*required*for*repairing*the*chitin*septum*during*cytokinesis;*
transcription*activated*by*mating*factor"
N UHOR_00740*,U
HOR_00723*,UH
OR_06435*
000E+0 sr15181*,sr1
0158*,sr1013
9*
000E+0
pf01387 um10312* 1EF39 um10312 probable*small*nuclear*ribonucleoprotein*E other SME1 YOR159C "Core*Sm*protein*Sm*E;*part*of*heteroheptameric*complex*(with*Smb1p,*Smd1p,*Smd2p,*Smd3p,*Smx3p,*
and*Smx2p)*that*is*part*of*the*spliceosomal*U1,*U2,*U4,*and*U5*snRNPs;*homolog*of*human*Sm*E"
N UHOR_04279* 10EF39 sr13727* 20EF39
pf05771 um10533* 000E+0 um10533 kgd1*probable*KGD1*F*alphaFketoglutarate*
dehydrogenase
other KGD1 "YIL125W,*SPBC3H7.03c" "Component*of*the*mitochondrial*alphaFketoglutarate*dehydrogenase*complex,*which*catalyzes*a*key*
step*in*the*tricarboxylic*acid*(TCA)*cycle,*the*oxidative*decarboxylation*of*alphaFketoglutarate*to*form*
succinylFCoA"
N UHOR_06953* 000E+0 sr15339* 000E+0
pf04432 um10682* 000E+0 um10682 cyb2*probable*CYB2*F*LFlactate*dehydrogenase*
(cytochrome*b2)
other CYB2 "YML054C,*SPAPB1A11.03" "Cytochrome*b2*(LFlactate*cytochromeFc*oxidoreductase),*component*of*the*mitochondrial*
intermembrane*space,*required*for*lactate*utilization;*expression*is*repressed*by*glucose*and*anaerobic*
conditions"
N UHOR_01415* 000E+0 sr15621* 000E+0
pf02796 um11243* 2EF75 um11243 related*to*ISA1*F*Fe/S*cluster*assembly*protein other ISA1 "YLL027W,*SPCC645.03c" "Mitochondrial*matrix*protein*involved*in*biogenesis*of*the*ironFsulfur*(Fe/S)*cluster*of*Fe/S*proteins,*
isa1*deletion*causes*loss*of*mitochondrial*DNA*and*respiratory*deficiency;*depletion*reduces*growth*on*
nonfermentable*carbon*sources"
N UHOR_01250* 3EF75 sr12121* 300EF75
pf02330 um11284* 000E+0 um11284 related*to*glycogeninF2*beta other GLG2 "YJL137C,*SPBC4C3.08" "SelfFglucosylating*initiator*of*glycogen*synthesis,*also*glucosylates*nFdodecylFbetaFDFmaltoside;*similar*
to*mammalian*glycogenin"
N UHOR_01677* 000E+0 sr12408* 000E+0
pf03560 um11363* 6EF132 um11363 "related*to*RIB5*F*riboflavin*synthase,*alpha*chain" other RIB5 "YBR256C,*SPCC1450.13c" Riboflavin*synthase;*catalyzes*the*last*step*of*the*riboflavin*biosynthesis*pathway N UHOR_02542* 60EF135 sr12782* 80EF135
pf00651 um11427* 10EF66 um11427 conserved*hypothetical*protein other SYF2 "YGR129W,*SPBC3E7.13c" Component*of*the*spliceosome*complex*involved*in*preFmRNA*splicing;*involved*in*regulation*of*cell*
cycle*progression
N UHOR_00369* 80EF60 sr11586* 40EF87
pf02046 um11638* 3EF63 um11638 related*to*glucosamine*6Fphosphate*nFacetyltransferase other GNA1 "YFL017C,*SPAC16E8.03" "Evolutionarily*conserved*glucosamineF6Fphosphate*acetyltransferase*required*for*multiple*cell*cycle*
events*including*passage*through*START,*DNA*synthesis,*and*mitosis;*involved*in*UDPFNF
acetylglucosamine*synthesis,*forms*GlcNAc6P*from*AcCoA"
N UHOR_04550* 10EF63 sr13939* 20EF60
pf00997 um11662* 5EF144 um11662 related*to*ECM4*F*involved*in*cell*wall*biogenesis*and*
architecture
other ECM4 "YKR076W,*SPCC1281.07c" Omega*class*glutathione*transferase;*not*essential;*similar*to*Ygr154cp;*green*fluorescent*protein*(GFP)F
fusion*protein*localizes*to*the*cytoplasm
N UHOR_02186* 90EF144 sr12531* 50EF141
pf03779 um11986* 000E+0 um11986 Kin4*cFterminal*kinesin other KAR3 YPR141C "MinusFendFdirected*microtubule*motor*that*functions*in*mitosis*and*meiosis,*localizes*to*the*spindle*
pole*body*and*localization*is*dependent*on*functional*Cik1p,*required*for*nuclear*fusion*during*mating;*
potential*Cdc28p*substrate"
N UHOR_06484* 000E+0 sr15392* 000E+0
pf05634 um12177* 000E+0 um12177 xks1*probable*XKS1*F*xylulokinase other XKS1 "YGR194C,*SPCPJ732.02c" "Xylulokinase,*converts*DFxylulose*and*ATP*to*xylulose*5Fphosphate*and*ADP;*rate*limiting*step*in*
fermentation*of*xylulose;*required*for*xylose*fermentation*by*recombinant*S.*cerevisiae*strains"
N UHOR_02849* 000E+0 sr10654* 000E+0
pf03622 um12183* 000E+0 um12183 conserved*hypothetical*protein other PIB1 "YDR313C,*SPBC36B7.05c" "RINGFtype*ubiquitin*ligase*of*the*endosomal*and*vacuolar*membranes,*binds*phosphatidylinositol(3)F
phosphate;*contains*a*FYVE*finger*domain"
N UHOR_03485* 000E+0 sr13323* 000E+0
pf06567 um12209* 000E+0 um12209 related*to*SMC4*F*Stable*Maintenance*of*Chromosomes other CSM1 "YCR086W,*NCU09063.2" "Nucleolar*protein*that*forms*a*complex*with*Lrs4p*which*binds*Mam1p*at*kinetochores*during*meiosis*I*
to*mediate*accurate*chromosome*segregation,*may*be*involved*in*premeiotic*DNA*replication;*possible*
role*in*telomere*maintenance"
Y UHOR_04895* 000E+0 sr16457 300EF48
pf04311 um12284* 2EF81 um12284 probable*nuclear*cap*binding*protein*subunit*2 other CBC2 "YPL178W,*SPBC13A2.01c" "Small*subunit*of*the*heterodimeric*cap*binding*complex*that*also*contains*Sto1p,*component*of*the*
spliceosomal*commitment*complex;*interacts*with*Npl3p,*possibly*to*package*mRNA*for*export*from*the*
nucleus;*contains*an*RNAFbinding*motif"
Y UHOR_07761* 100EF90 sr15739* 80EF87
pf00580 um15009* 30EF156 um15009 conserved*hypothetical*protein other GAC1 YOR178C "Regulatory*subunit*for*Glc7p*typeF1*protein*phosphatase*(PP1),*tethers*Glc7p*to*Gsy2p*glycogen*
synthase,*binds*Hsf1p*heat*shock*transcription*factor,*required*for*induction*of*some*HSFFregulated*
genes*under*heat*shock"
N UHOR_00973* 000E+0 sr11907* 000E+0
pf02642 um15043.2 10EF96 um15043.2 probable*ENA2*F*Plasma*membrane*PFtype*ATPase other ENA2 "YDR039C,*SPBC839.06" "PFtype*ATPase*sodium*pump,*involved*in*Na+*efflux*to*allow*salt*tolerance;*likely*not*involved*in*Li+*
efflux"
N UHOR_02598 800EF102 sr12820 3EF99
pf00514 um15043.2*,u
m00204*
000E+0 um15043.2 probable*ENA2*F*Plasma*membrane*PFtype*ATPase other ENA2 "YDR039C,*SPBC839.06" "PFtype*ATPase*sodium*pump,*involved*in*Na+*efflux*to*allow*salt*tolerance;*likely*not*involved*in*Li+*
efflux"
N UHOR_02598*,U
HOR_00318*
000E+0 sr11550*,sr1
2820*
000E+0
pf04558 um15043.2*,u
m00204*
000E+0 um15043.2 probable*ENA2*F*Plasma*membrane*PFtype*ATPase other ENA2 "YDR039C,*SPBC839.06" "PFtype*ATPase*sodium*pump,*involved*in*Na+*efflux*to*allow*salt*tolerance;*likely*not*involved*in*Li+*
efflux"
N UHOR_02598*,U
HOR_00318*
000E+0 sr11550*,sr1
2820*
000E+0
pf00800 um15070* 000E+0 um15070 probable*to*GDP/GTP*exchange*factor*Rom2p other ROM2 "YLR371W,*SPAC1006.06" "GDP/GTP*exchange*protein*(GEP)*for*Rho1p*and*Rho2p;*mutations*are*synthetically*lethal*with*
mutations*in*rom1,*which*also*encodes*a*GEP"
N UHOR_05941* 000E+0 sr14815* 000E+0
-
pf05965 um03924 40EF12 um03924 Rep1,*repellent*protein*1*precursor pathogenicity? N UHOR_05948 1EF12 sr14829 2EF12pf02509 um05543 40EF81 um05543 Don3,*Ste20Flike*kinase*triggers*septin*reorganization*
during*secondary*septum*formationpathogenicity? UHOR_03730* 000E+0 sr13462* 000E+0
pf01924 um02874* 000E+0 um02874 related*to*SGS1*F*DNA*helicase Regulation*of*crossover*frequency SGS1 YMR190C "Nucleolar*DNA*helicase*of*the*RecQ*family*involved*in*maintenance*of*genome*integrity,*regulates*chromosome*synapsis*and*meiotic*crossing*over;*has*similarity*to*human*BLM*and*WRN*helicases*implicated*in*Bloom*and*Werner*syndromes"
N UHOR_04532* 000E+0 sr13928* 2EF174
pf02269 um03481* 20EF9 um03481 related*to*MLH3*F*insertion*and*deletion*mismatch*repair*protein
Regulation*of*crossover*frequency MLH3 YPL164C Protein*involved*in*DNA*mismatch*repair*and*crossingFover*during*meiotic*recombination;*forms*a*complex*with*Mlh1p;*mammalian*homolog*is*implicated*mammalian*microsatellite*instability
Y UHOR_05356* 90EF15 sr14470* 9EF12
pf03077 um05208* 000E+0 um05208 related*to*MLH1*F*DNA*mismatch*repair*protein Regulation*of*crossover*frequency MLH1 "YMR167W,*NCU08309.1" Protein*required*for*mismatch*repair*in*mitosis*and*meiosis*as*well*as*crossing*over*during*meiosis;*forms*a*complex*with*Pms1p*and*Msh2pFMsh3p*during*mismatch*repair;*human*homolog*is*associated*with*hereditary*nonFpolyposis*colon*cancer
N UHOR_03183* 000E+0 sr13114* 000E+0
pf06233 um12155.2 300EF51 um12155.2 related*to*MutS*protein*homolog*5 Regulation*of*crossover*frequency MSH5 "YDL154W,*NCU09384.2" "Protein*of*the*MutS*family,*forms*a*dimer*with*Msh4p*that*facilitates*crossovers*between*homologs*during*meiosis;*msh5FY823H*mutation*confers*tolerance*to*DNA*alkylating*agents;*homologs*present*in*C.*elegans*and*humans"
Y UHOR_00738 200EF51 sr10153.2 50EF45
pf06232 um12155.2* 8EF81 um12155.2 related*to*MutS*protein*homolog*5 Regulation*of*crossover*frequency MSH5 "YDL154W,*NCU09384.2" "Protein*of*the*MutS*family,*forms*a*dimer*with*Msh4p*that*facilitates*crossovers*between*homologs*during*meiosis;*msh5FY823H*mutation*confers*tolerance*to*DNA*alkylating*agents;*homologs*present*in*C.*elegans*and*humans"
Y UHOR_00738* 2EF78 sr10153.2* 400EF75
pf02394 um01085* 000E+0 um01085 rad50*probable*RAD50*F*DNA*repair*protein Removal*of*Spo11 RAD50 "YNL250W,*NCU00901.1" "Subunit*of*MRX*complex,*with*Mre11p*and*Xrs2p,*involved*in*processing*doubleFstrand*DNA*breaks*in*vegetative*cells,*initiation*of*meiotic*DSBs,*telomere*maintenance,*and*nonhomologous*end*joining"
N UHOR_01639* 000E+0 sr12383* 000E+0
pf04420 um04704* 000E+0 um04704 related*to*MRE11*F*DNA*repair*and*meiotic*recombination*protein
Removal*of*Spo11 MRE11 "YMR224C,*NCU08730.2" "Subunit*of*a*complex*with*Rad50p*and*Xrs2p*(RMX*complex)*that*functions*in*repair*of*DNA*doubleFstrand*breaks*and*in*telomere*stability,*exhibits*nuclease*activity*that*appears*to*be*required*for*RMX*function;*widely*conserved"
Y UHOR_07541* 000E+0 sr15581* 000E+0
pf00361 um01857* 90EF114 um01857 conserved*hypothetical*protein Resolution*of*recombination*intermediates SLX1 "YBR228W,*NCU01236.1" "Subunit*of*a*complex,*with*Slx4p,*that*hydrolyzes*5'*branches*from*duplex*DNA*in*response*to*stalled*or*converging*replication*forks;*function*overlaps*with*that*of*Sgs1pFTop3p"
N UHOR_02758* 200EF120 sr10553.2* 5EF108
pf05520 um03276* 100EF9 um03276 related*to*SRP40*F*serineFrich*protein*with*a*role*in*preFribosome*assembly*or*transport
Resolution*of*recombination*intermediates MMS4 "YBR098W,*NCU04047.1" Subunit*of*the*structureFspecific*Mms4pFMus81p*endonuclease*that*cleaves*branched*DNA;*involved*in*recombination*and*DNA*repair
N no*hit no*hit sr14276 3EF3
pf02287 um03501* 000E+0 um03501 probable*DNA*topoisomerase*II Resolution*of*recombination*intermediates TOP2 "YNL088W,*NCU06338.1" "Essential*type*II*topoisomerase,*relieves*torsional*strain*in*DNA*by*cleaving*and*reFsealing*the*phosphodiester*backbone*of*both*positively*and*negatively*supercoiled*DNA;*cleaves*complementary*strands;*localizes*to*axial*cores*in*meiosis"
N UHOR_05386* 000E+0 sr14492* 000E+0
pf01421 um04630* 200EF96 um04630 related*to*MUS81*F*endonuclease*involved*in*DNA*repair*and*replication*fork*stability
Resolution*of*recombination*intermediates MUS81 "YDR386W,*NCU07457.1" "HelixFhairpinFhelix*protein,*involved*in*DNA*repair*and*replication*fork*stability;*functions*as*an*endonuclease*in*complex*with*Mms4p;*interacts*with*Rad54p"
N UHOR_06657* 30EF93 sr15514* 100EF96
pf03209 um05101* 000E+0 um05101 Top1*DNA*topoisomerase*I Resolution*of*recombination*intermediates TOP1 "YOL006C,*NCU09118.1" "Topoisomerase*I,*nuclear*enzyme*that*relieves*torsional*strain*in*DNA*by*cleaving*and*reFsealing*the*phosphodiester*backbone;*relaxes*both*positively*and*negatively*supercoiled*DNA;*functions*in*replication,*transcription,*and*recombination"
N UHOR_03033* 000E+0 sr13004* 000E+0
pf03930 um11929* 000E+0 um11929 related*to*DNA*topoisomerase*III*alpha Resolution*of*recombination*intermediates TOP3 "YLR234W,*NCU00081.1" "DNA*Topoisomerase*III,*conserved*protein*that*functions*in*a*complex*with*Sgs1p*and*Rmi1p*to*relax*singleFstranded*negativelyFsupercoiled*DNA*preferentially,*involved*in*telomere*stability*and*regulation*of*mitotic*recombination"
N UHOR_04384* 000E+0 sr13804* 000E+0
pf02532 um03599* 000E+0 um03599 cdc12*probable*CDC12*F*septin Septins SPR3 "YGR059W,*SPCC188.12,*NCU03795.2" SporulationFspecific*homolog*of*the*yeast*CDC3/10/11/12*family*of*bud*neck*microfilament*genes;*septin*protein*involved*in*sporulation;*regulated*by*ABFI
Y UHOR_05523* 000E+0 sr11143.2* 000E+0
pf01166 um10503* 000E+0 um10503 probable*cell*division*control*protein*CDC3 Septins CDC3 "YLR314C,*SPAC24C9.15c" "Component*of*the*septin*ring*of*the*motherFbud*neck*that*is*required*for*cytokinesis;*septins*recruit*proteins*to*the*neck*and*can*act*as*a*barrier*to*diffusion*at*the*membrane,*and*they*comprise*the*10nm*filaments*seen*with*EM"
N UHOR_05696* 000E+0 sr11206* 000E+0
pf06579 um10644* 2EF162 um10644 cdc10*probable*CDC10*F*septin Septins CDC10 "YCR002C,*SPAC821.06" "Component*of*the*septin*ring*of*the*motherFbud*neck*that*is*required*for*cytokinesis;*septins*recruit*proteins*to*the*neck*and*can*act*as*a*barrier*to*diffusion*at*the*membrane,*and*they*comprise*the*10nm*filaments*seen*with*EM"
N UHOR_05011* 200EF165 sr11123* 50EF165
pf06115 um00560* 000E+0 um00560 probable*glycogen*synthase*kinase*3*alpha Signal*transduction RIM11 "YMR139W,*NCU04185.2" Protein*kinase*required*for*signal*transduction*during*entry*into*meiosis;*promotes*the*formation*of*the*Ime1pFUme6p*complex*by*phosphorylating*Ime1p*and*Ume6p;*shares*similarity*with*mammalian*glycogen*synthase*kinase*3Fbeta
Y UHOR_00864* 000E+0 sr11838* 000E+0
pf01047 um00986* 100EF114 um00986 Ras1*small*GFprotein*Ras1 Signal*transduction RAS2 "YNL098C,*SPAC17H9.09c" "GTPFbinding*protein*that*regulates*the*nitrogen*starvation*response,*sporulation,*and*filamentous*growth;*farnesylation*and*palmitoylation*required*for*activity*and*localization*to*plasma*membrane;*homolog*of*mammalian*Ras*protoFoncogenes"
N UHOR_01498* 2EF105 sr12287* 5EF105
pf01333 um02902* 300EF156 um02902 related*to*BAG7*F*Rho*GTPase*activating*protein Signal*transduction BAG7 "YOR134W,*SPBC17F3.01c*(SPBC557.01)" "Rho*GTPase*activating*protein*(RhoGAP),*stimulates*the*intrinsic*GTPase*activity*of*Rho1p,*which*plays*a*role*in*actin*cytoskeleton*organization*and*control*of*cell*wall*synthesis;*structurally*and*functionally*related*to*Sac7p"
N UHOR_04579* 80EF156 sr13961* 4EF156
pf05073 um02989* 50EF126 um02989 related*to*IME4*F*positive*transcription*factor*for*IME2 Signal*transduction IME4 YGL192W Probable*mRNA*N6Fadenosine*methyltransferase*that*is*required*for*IME1*transcript*accumulation*and*for*sporulation;*expression*is*induced*in*starved*MATa/MAT*alpha*diploid*cells
Y UHOR_04696* 3EF123 sr14049* 20EF126
pf02902 um03080* 000E+0 um03080 probable*serine/threonine*protein*phosphatase*PP1 Signal*transduction GLC7 YER133W "Catalytic*subunit*of*type*1*serine/threonine*protein*phosphatase,*involved*in*many*processes*including*glycogen*metabolism,*sporulation,*and*mitosis;*interacts*with*multiple*regulatory*subunits;*predominantly*isolated*with*Sds22p"
N UHOR_04823* 000E+0 sr14139* 000E+0
pf06580 um03216* 000E+0 um03216 tor1*probable*TOR1*F*1Fphosphatidylinositol*3Fkinase Signal*transduction TOR1 YJR066W "PIKFrelated*protein*kinase*and*rapamycin*target;*subunit*of*TORC1,*a*complex*that*controls*growth*in*response*to*nutrients*by*regulating*translation,*transcription,*ribosome*biogenesis,*nutrient*transport*and*autophagy;*involved*in*meiosis"
N UHOR_05010* 000E+0 sr11121* 000E+0
pf05212 um05453* 700EF90 um05453 related*to*Tyrosine*specific*protein*phosphatase*and*dual*specificity*protein*phosphatase
Signal*transduction YVH1 YIR026C "Protein*phosphatase*involved*in*vegetative*growth*at*low*temperatures,*sporulation,*and*glycogen*accumulation;*transcription*induced*by*low*temperature*and*nitrogen*starvation;*member*of*the*dualFspecificity*family*of*protein*phosphatases"
N UHOR_07822* 700EF159 sr16137* 300EF117
pf05109 um05544* 000E+0 um05544 probable*calmodulinFdependent*protein*kinase*type*1 Signal*transduction CMK2 "YOL016C,*SPACUNK12.02c" "CalmodulinFdependent*protein*kinase;*may*play*a*role*in*stress*response,*many*CA++/calmodulan*dependent*phosphorylation*substrates*demonstrated*in*vitro,*amino*acid*sequence*similar*to*Cmk1p*and*mammalian*Cam*Kinase*II"
N UHOR_07949* 000E+0 sr16226* 000E+0
pf02149 um06019* 20EF171 um06019 cmk1*probable*CMK1*F*Ca2+/calmodulinFdependent*ser/thr*protein*kinase*type*I
Signal*transduction MEK1 "YOR351C,*NCU09123.2" "MeiosisFspecific*serine/threonine*protein*kinase,*functions*in*meiotic*checkpoint,*promotes*recombination*between*homologous*chromosomes*by*suppressing*double*strand*break*repair*between*sister*chromatids"
Y UHOR_08553* 20EF174 sr16646* 60EF171
pf06644 um06212* 2EF81 um06212 "related*to*SSN8*F*DNAFdirected*RNA*polymerase*II*holoenzyme*and*SRB*subcomplex*subunit,*cyclin*C*homolog"
Signal*transduction SSN8 YNL025C "CyclinFlike*component*of*the*RNA*polymerase*II*holoenzyme,*involved*in*phosphorylation*of*the*RNA*polymerase*II*CFterminal*domain;*involved*in*glucose*repression*and*telomere*maintenance"
N UHOR_08803* 10EF75 sr11334* 70EF93
pf03050 um06337* 3EF147 um06337 Wee1*wee1*kinase Signal*transduction SWE1 YJL187C Protein*kinase*that*regulates*the*G2/M*transition*by*inhibition*of*Cdc28p*kinase*activity;*localizes*to*the*nucleus*and*to*the*daughter*side*of*the*motherFbud*neck;*homolog*of*S.*pombe*Wee1p;*potential*Cdc28p*substrate
N UHOR_03333* 000E+0 sr15950 4EF24
pf03617 um10056* 000E+0 um10056 related*to*serine/threonineFprotein*kinase Signal*transduction RCK2 YLR248W Protein*kinase*involved*in*the*response*to*oxidative*and*osmotic*stress;*identified*as*suppressor*of*S.*pombe*cell*cycle*checkpoint*mutations
N UHOR_03516* 000E+0 sr13347* 000E+0
pf06284 um10107* 000E+0 um10107 probable*mitogenFactivated*protein*kinase*MpkA Signal*transduction SMK1 "YPR054W,*NCU09842.2" Middle*sporulationFspecific*mitogenFactivated*protein*kinase*(MAPK)*required*for*production*of*the*outer*spore*wall*layers;*negatively*regulates*activity*of*the*glucan*synthase*subunit*Gsc2p
Y UHOR_00662* 000E+0 sr11786* 000E+0
pf04469 um10496 4EF57 um10496 related*to*KIC1*F*ser/thr*protein*kinase*that*interacts*with*Cdc31p
Signal*transduction SPS1 "YDR523C,*NCU00772.2" "Putative*protein*serine/threonine*kinase*expressed*at*the*end*of*meiosis*and*localized*to*the*prospore*membrane,*required*for*correct*localization*of*enzymes*involved*in*spore*wall*synthesis"
Y UHOR_07746* 000E+0 sr15729* 000E+0
pf01144 um10496* 000E+0 um10496 related*to*KIC1*F*ser/thr*protein*kinase*that*interacts*with*Cdc31p
Signal*transduction SPS1 "YDR523C,*NCU00772.2" "Putative*protein*serine/threonine*kinase*expressed*at*the*end*of*meiosis*and*localized*to*the*prospore*membrane,*required*for*correct*localization*of*enzymes*involved*in*spore*wall*synthesis"
Y UHOR_05679* 000E+0 sr11189* 000E+0
pf05329 um10705* 300EF153 um10705 Cdk1*cyclinFdependent*kinase*1 Signal*transduction CDC28 YBR160W Catalytic*subunit*of*the*main*cell*cycle*cyclinFdependent*kinase*(CDK);*alternately*associates*with*G1*cyclins*(CLNs)*and*G2/M*cyclins*(CLBs)*which*direct*the*CDK*to*specific*substrates
N UHOR_08118* 200EF156 sr10045* 4EF156
pf05841 um11410* 000E+0 um11410 Crk1*cdkFrelated*kinase*1 Signal*transduction IME2 "YJL106W,*NCU01498.2" "Serine/threonine*protein*kinase*involved*in*activation*of*meiosis,*associates*with*Ime1p*and*mediates*its*stability,*activates*Ndt80p;*IME2*expression*is*positively*regulated*by*Ime1p"
Y UHOR_04041* 000E+0 sr10962* 000E+0
pf05914 um11677.2* 000E+0 um11677.2 related*to*serine/threonine*protein*kinase Signal*transduction RIM15 "YFL033C,*NCU07378.2" "GlucoseFrepressible*protein*kinase*involved*in*signal*transduction*during*cell*proliferation*in*response*to*nutrients,*specifically*the*establishment*of*stationary*phase;*identified*as*a*regulator*of*IME2;*substrate*of*Pho80pFPho85p*kinase"
Y UHOR_02215* 000E+0 sr12574* 000E+0
pf06569 um12208* 10EF174 um12208 related*to*SSN3*F*cyclinFdependent*CTD*kinase Signal*transduction SSN3 YPL042C "CyclinFdependent*protein*kinase,*component*of*RNA*polymerase*II*holoenzyme;*involved*in*phosphorylation*of*the*RNA*polymerase*II*CFterminal*domain;*involved*in*glucose*repression"
N UHOR_04893* 8EF159 sr14186* 70EF168
pf01050 um00996.2* 000E+0 um00996.2 conserved*hypothetical*protein Spore*wall*formation SPO73 "YER046W,*SPAC1296.04" "MeiosisFspecific*protein*of*unknown*function,*required*for*spore*wall*formation*during*sporulation;*dispensible*for*both*nuclear*divisions*during*meiosis"
Y UHOR_01516* 000E+0 sr12297* 000E+0
pf04302 um02019* 8EF159 um02019 probable*Chitin*deacetylase Spore*wall*formation CDA2 YLR308W "Chitin*deacetylase,*together*with*Cda1p*involved*in*the*biosynthesis*ascospore*wall*component,*chitosan;*required*for*proper*rigidity*of*the*ascospore*wall"
Y UHOR_03000* 5EF150 sr12981* 20EF153
pf04976 um02889* 60EF132 um02889 related*to*SEC9*F*protein*transport*protein Spore*wall*formation SPO20 "YMR017W,*NCU09243.2" "MeiosisFspecific*subunit*of*the*tFSNARE*complex,*required*for*prospore*membrane*formation*during*sporulation;*similar*to*but*not*functionally*redundant*with*Sec9p;*SNAPF25*homolog"
Y UHOR_04554* 4EF129 sr13943* 20EF126
pf02924 um04961* 000E+0 um04961 conserved*hypothetical*protein Spore*wall*formation SPO71 YDR104C "MeiosisFspecific*protein*of*unknown*function,*required*for*spore*wall*formation*during*sporulation;*dispensable*for*both*nuclear*divisions*during*meiosis"
Y UHOR_07098* 000E+0 sr15841* 000E+0
pf04606 um05664* 000E+0 um05664 related*to*Cytochrome*P450 Spore*wall*formation DIT2 YDR402C "NFformyltyrosine*oxidase,*sporulationFspecific*microsomal*enzyme*involved*in*the*production*of*N,NFbisformyl*dityrosine*required*for*spore*wall*maturation,*homologous*to*cytochrome*PF450s"
Y UHOR_07421* 000E+0 sr15988* 000E+0
pf00506 um00196* 30EF114 um00196 fun34*probable*FUN34*F*transmembrane*protein*involved*in*ammonia*production
Sporulation ADY2 "YCR010C,*NCU06043.2" Acetate*transporter*required*for*normal*sporulation;*phosphorylated*in*mitochondria Y UHOR_00308* 2EF111 sr11542* 9EF114
pf00984 um00370 30EF90 um00370 spo14*probable*SPO14*F*phospholipase*D Sporulation SPO14 "YKR031C,*NCU03955.2" "Phospholipase*D,*catalyzes*the*hydrolysis*of*phosphatidylcholine,*producing*choline*and*phosphatidic*acid;*involved*in*Sec14pFindependent*secretion;*required*for*meiosis*and*spore*formation;*differently*regulated*in*secretion*and*meiosis"
Y UHOR_00572 6EF90 sr11726 200EF90
-
pf03521 um00370* 000E+0 um00370 spo14*probable*SPO14*F*phospholipase*D Sporulation SPO14 "YKR031C,*NCU03955.2" "Phospholipase*D,*catalyzes*the*hydrolysis*of*phosphatidylcholine,*producing*choline*and*phosphatidic*acid;*involved*in*Sec14pFindependent*secretion;*required*for*meiosis*and*spore*formation;*differently*regulated*in*secretion*and*meiosis"
Y UHOR_00572* 000E+0 sr11726* 000E+0
pf02067 um00532* 6EF78 um00532 putative*protein Sporulation DON1 YDR273W "MeiosisFspecific*component*of*the*spindle*pole*body,*part*of*the*leading*edge*protein*(LEP)*coat,*forms*a*ringFlike*structure*at*the*leading*edge*of*the*prospore*membrane*during*meiosis*II"
Y UHOR_03926* 7EF87 sr11808* 200EF81
pf06732 um04154* 5EF45 um04154 conserved*hypothetical*protein Sporulation SPO7 YAL009W "Regulatory*subunit*of*Nem1pFSpo7p*phosphatase*holoenzyme,*which*regulates*nuclear*growth*by*controlling*recruitment*of*Pah1p*onto*promoters*of*phospholipid*biosynthetic*genes;*required*for*normal*nuclear*envelope*morphology*and*sporulation"
N UHOR_06231* 20EF72 sr15034* 40EF66
pf05654 um05550* 5EF177 um05550 Sporulation SPR1 "YOR190W,*NCU03914.2" "SporulationFspecific*exoF1,3FbetaFglucanase;*contributes*to*ascospore*thermoresistance" Y UHOR_06823* 300EF171 sr15250* 20EF159pf06434 um11104* 000E+0 um11104 Nir1*putative*nitrite*reductase Sporulation ADY4 YLR227C "Structural*component*of*the*meiotic*outer*plaque,*which*is*a*membraneForganizing*center*that*
assembles*on*the*cytoplasmic*face*of*the*spindle*pole*body*during*meiosis*II*and*triggers*the*formation*of*the*prospore*membrane"
Y UHOR_05849* 000E+0 sr14750* 000E+0
pf00819 um11266* 000E+0 um11266 probable*lysophospholipase*(lpl) Sporulation SPO1 "YNL012W,*NCU03141.2" "MeiosisFspecific*protein*with*similarity*to*phospholipase*B,*required*for*meiotic*spindle*pole*body*duplication*and*separation;*required*for*spore*formation"
Y UHOR_01564* 000E+0 sr12339* 000E+0
pf00591 um02083* 000E+0 um02083 rad54*probable*RAD54*F*DNAFdependent*ATPase*of*the*Snf2p*family
Strand*invasion RAD54 "YGL163C,*NCU02348.1" "DNAFdependent*ATPase,*stimulates*strand*exchange*by*modifying*the*topology*of*doubleFstranded*DNA;*involved*in*the*recombinational*repair*of*doubleFstrand*breaks*in*DNA*during*vegetative*growth*and*meiosis;*member*of*the*SWI/SNF*family"
N UHOR_03459* 000E+0 sr13306* 000E+0
pf01970 um02579* 6EF81 um02579 "related*to*RFA2*F*DNA*replication*factor*A,*36*kDa*subunit"
Strand*invasion RFA2 "YNL312W,*NCU07717.1" "Subunit*of*heterotrimeric*Replication*Protein*A*(RPA),*which*is*a*highly*conserved*singleFstranded*DNA*binding*protein*involved*in*DNA*replication,*repair,*and*recombination"
N UHOR_04124* 600EF87 sr13642* 100EF90
pf06516 um03290* 000E+0 um03290 Rad51*DNA*repair*protein*RAD51 Strand*invasion RAD51 "YER095W,*NCU02741.1" "Strand*exchange*protein,*forms*a*helical*filament*with*DNA*that*searches*for*homology;*involved*in*the*recombinational*repair*of*doubleFstrand*breaks*in*DNA*during*vegetative*growth*and*meiosis;*homolog*of*Dmc1p*and*bacterial*RecA*protein"
N UHOR_05125* 000E+0 sr14292* 000E+0
pf01252 um03676* 000E+0 um03676 related*to*RAD54*F*DNAFdependent*ATPase*of*the*Snf2p*family
Strand*invasion RDH54 YBR073W "DNAFdependent*ATPase,*stimulates*strand*exchange*by*modifying*the*topology*of*doubleFstranded*DNA;*involved*in*recombinational*repair*of*DNA*doubleFstrand*breaks*during*mitosis*and*meiosis;*proposed*to*be*involved*in*crossover*interference"
N UHOR_05627* 000E+0 sr14625* 000E+0
pf01623 um04989* 300EF132 um04989 related*to*RAD52*F*recombination*and*DNA*repair*protein
Strand*invasion RAD52 "YML032C,*NCU04275.1" Protein*that*stimulates*strand*exchange*by*facilitating*Rad51p*binding*to*singleFstranded*DNA;*anneals*complementary*singleFstranded*DNA;*involved*in*the*repair*of*doubleFstrand*breaks*in*DNA*during*vegetative*growth*and*meiosis
N UHOR_07140* 4EF18 sr15872* 200EF129
pf03095 um05156* 000E+0 um05156 probable*Replication*factorFA*protein*1 Strand*invasion RFA1 "YAR007C,*NCU03606.1" "Subunit*of*heterotrimeric*Replication*Protein*A*(RPA),*which*is*a*highly*conserved*singleFstranded*DNA*binding*protein*involved*in*DNA*replication,*repair,*and*recombination"
N UHOR_03102* 000E+0 sr13061* 000E+0
pf03252 um00766* 1EF45 um00766 putative*protein Synaptonemal*complex ZIP1 "YDR285W,*NCU00658.2" Transverse*filament*protein*of*the*synaptonemal*complex;*required*for*normal*levels*of*meiotic*recombination*and*pairing*between*homologous*chromosome*during*meiosis;*potential*Cdc28p*substrate
Y UHOR_01172* 100EF51 sr12058* 1EF45
pf03283 um00592* 000E+0 um00592 related*to*Nuclear*receptor*coFrepressor/HDAC3*complex*subunit*TBLR1
Transcription*factor*/*Gene*regulation SIF2 YBR103W "WD40*repeatFcontaining*subunit*of*the*Set3C*histone*deacetylase*complex,*which*represses*early/middle*sporulation*genes;*antagonizes*telomeric*silencing;*binds*specifically*to*the*Sir4p*NFterminus"
N UHOR_00919* 000E+0 sr11870* 000E+0
pf05822 um01961* 2EF9 um01961 hypothetical*protein Transcription*factor*/*Gene*regulation UME6 YDR207C "Key*transcriptional*regulator*of*early*meiotic*genes,*binds*URS1*upstream*regulatory*sequence,*couples*metabolic*responses*to*nutritional*cues*with*initiation*and*progression*of*meiosis,*forms*complex*with*Ime1p,*and*also*with*Sin3pFRpd3p"
N UHOR_02913* 900EF12 sr10717* 1EF9
pf01537 um02405* 700EF156 um02405 conserved*hypothetical*protein Transcription*factor*/*Gene*regulation KAR4 YCL055W "Transcription*factor*required*for*gene*regulation*in*repsonse*to*pheromones;*also*required*during*meiosis;*exists*in*two*forms,*a*slowerFmigrating*form*more*abundant*during*vegetative*growth*and*a*fasterFmigrating*form*induced*by*pheromone"
N UHOR_03945* 4EF159 sr10812* 100EF153
pf03909 um02775* 1EF27 um02775 hypothetical*protein Transcription*factor*/*Gene*regulation NDT80 "YHR124W,*NCU09915.2" MeiosisFspecific*transcription*factor*required*for*exit*from*pachytene*and*for*full*meiotic*recombination;*activates*middle*sporulation*genes;*competes*with*Sum1p*for*binding*to*promoters*containing*middle*sporulation*elements*(MSE)
Y UHOR_04421* 3EF45 sr13828* 70EF30
pf05333 um05338* 000E+0 um05338 related*to*transcription*factor*MBP1 Transcription*factor*/*Gene*regulation SWI6 "YLR182W,*SPBC336.12c" "Transcription*cofactor,*forms*complexes*with*DNAFbinding*proteins*Swi4p*and*Mbp1p*to*regulate*transcription*at*the*G1/S*transition;*involved*in*meiotic*gene*expression;*localization*regulated*by*phosphorylation;*potential*Cdc28p*substrate"
N UHOR_08158* 30EF78 sr16322* 000E+0
pf05108 um05545* 000E+0 um05545 related*to*proteinFlysine*NFmethyltransferase Transcription*factor*/*Gene*regulation DOT1 YDR440W "Nucleosomal*histone*H3FLys79*methylase,*associates*with*transcriptionally*active*genes,*functions*in*gene*silencing*at*telomeres,*most*likely*by*directly*modulating*chromatin*structure*and*Sir*protein*localization"
N UHOR_07950* 000E+0 sr16227* 000E+0
pf02585 um05888* 90EF126 um05888 related*to*transcription*elongation*factor*TFIIS Transcription*factor*/*Gene*regulation DST1 YGL043W "General*transcription*elongation*factor*TFIIS,*enables*RNA*polymerase*II*to*read*through*blocks*to*elongation*by*stimulating*cleavage*of*nascent*transcripts*stalled*at*transcription*arrest*sites"
N UHOR_08387* 30EF120 sr16506* 20EF120
pf03305 um10426* 6EF174 um10426 PacC*transcription*factor*pacC Transcription*factor*/*Gene*regulation RIM101 YHL027W Transcriptional*repressor*involved*in*response*to*pH*and*in*cell*wall*construction;*required*for*alkaline*pHFstimulated*haploid*invasive*growth*and*sporulation;*activated*by*proteolytic*processing;*similar*to*A.*nidulans*PacC
N UHOR_06410* 000E+0 sr15166* 000E+0
pf05776 um10541* 200EF168 um10541 conserved*hypothetical*protein Transcription*factor*/*Gene*regulation RSV2*(S.*pombe)
SPBC1105.14 Transcription*factor*that*induces*stressFrelated*genes*during*spore*formation N UHOR_06975* 5EF60 sr15356* 600EF114
pf06760 um11222* 000E+0 um11222 "related*to*MBP1*F*transcription*factor,*subunit*of*the*MBF*factor"
Transcription*factor*/*Gene*regulation RES2*(S.*pombe)
SPAC22F3.09c Required*for*the*initiation*of*mitotic*and*premeiotic*DNA*synthesis.*Has*an*additional*role*in*meiotic*division.
N UHOR_08780* 000E+0 sr11315* 000E+0
pf03948 um11717 40EF42 um11717 related*to*SIN3*F*transcription*regulatory*protein Transcription*factor*/*Gene*regulation SIN3 YOL004W "Component*of*the*Sin3pFRpd3p*histone*deacetylase*complex,*involved*in*transcriptional*repression*and*activation*of*diverse*processes,*including*matingFtype*switching*and*meiosis;*involved*in*the*maintenance*of*chromosomal*integrity"
N UHOR_04320 60EF42 sr13761 60EF42
pf03947 um11717* 000E+0 um11717 related*to*SIN3*F*transcription*regulatory*protein Transcription*factor*/*Gene*regulation SIN3 YOL004W "Component*of*the*Sin3pFRpd3p*histone*deacetylase*complex,*involved*in*transcriptional*repression*and*activation*of*diverse*processes,*including*matingFtype*switching*and*meiosis;*involved*in*the*maintenance*of*chromosomal*integrity"
N UHOR_04320* 000E+0 sr13761* 000E+0
pf00498 um11828* 000E+0 um11828 hos2*probable*HOS2*F*putative*histone*deacetylase Transcription*factor*/*Gene*regulation HOS2 YGL194C "Histone*deacetylase*required*for*gene*activation*via*specific*deacetylation*of*lysines*in*H3*and*H4*histone*tails;*subunit*of*the*Set3*complex,*a*meioticFspecific*repressor*of*sporulation*specific*genes*that*contains*deacetylase*activity"
N UHOR_01015* 000E+0 sr11943* 000E+0
pf01077 um12055* 800EF42 um12055 related*to*ELC1*F*Elongin*C*transcription*elongation*factor*(CFterminal*fragment)
Transcription*factor*/*Gene*regulation ELC1 "YPL046C,*SPBC1861.07" "Elongin*C,*forms*heterodimer*with*Ela1p*that*participates*in*transcription*elongation;*required*for*ubiquitinFdependent*degradation*of*the*RNA*Polymerase*II*subunit*RPO21;*expression*dramatically*upregulated*during*sporulation"
N UHOR_01496* 5EF27 sr12278* 4EF39
pf03213 um12332* 60EF30 um12332 conserved*hypothetical*protein Transcription*factor*/*Gene*regulation YHP1 YDR451C "One*of*two*homeobox*transcriptional*repressors*(see*also*Yox1p),*that*bind*to*Mcm1p*and*to*early*cell*cycle*box*(ECB)*elements*of*cell*cycle*regulated*genes,*thereby*restricting*ECBFmediated*transcription*to*the*M/G1*interval"
N UHOR_03270* 80EF24 sr13171* 300EF27
pf01171 um15082* 4EF171 um15082 conserved*hypothetical*protein Transcription*factor*/*Gene*regulation SET3 YKR029C Defining*member*of*the*SET3*histone*deacetylase*complex*which*is*a*meiosisFspecific*repressor*of*sporulation*genes;*necessary*for*efficient*transcription*by*RNAPII;*one*of*two*yeast*proteins*that*contains*both*SET*and*PHD*domains
N UHOR_05704* 000E+0 sr11214* 000E+0
pf03479 um00581* 200EF69 um00581 conserved*hypothetical*protein Uncharacterized "ORF,*Uncharacterized"
"YOL019W,*SPCC1739.10" Protein*of*unknown*function;*green*fluorescent*protein*(GFP)Ffusion*protein*localizes*to*the*cell*periphery*and*vacuole
N UHOR_00899* 100EF75 sr11859* 30EF69
pf03464 um00735* 500EF138 um00735 conserved*hypothetical*protein Uncharacterized OXR1 "YPL196W,*SPAC8C9.16c" "Protein*of*unknown*function*required*for*normal*levels*of*resistance*to*oxidative*damage,*null*mutants*are*sensitive*to*hydrogen*peroxide;*member*of*a*conserved*family*of*proteins*found*in*eukaryotes*but*not*in*prokaryotes"
N UHOR_01130* 40EF177 sr12026* 1EF168
pf01281 um03761* 000E+0 um03761 related*to*aldehyde*dehydrogenase Uncharacterized MSC7 YHR039C "Protein*of*unknown*function,*green*fluorescent*protein*(GFP)Ffusion*protein*localizes*to*the*endoplasmic*reticulum;*msc7*mutants*are*defective*in*directing*meiotic*recombination*events*to*homologous*chromatids"
N UHOR_05727* 000E+0 sr14662* 000E+0
pf02131 um06092* 600EF171 um06092 conserved*hypothetical*protein Uncharacterized "ORF,*Uncharacterized"
"YNL115C,*SPAC23C11.06c" Putative*protein*of*unknown*function;*green*fluorescent*protein*(GFP)Ffusion*protein*localizes*to*mitochondria;*YNL115C*is*not*an*essential*gene
N UHOR_08645* 20EF168 sr16717* 60EF171
pf02070 um10302.2* 8EF36 um10302.2 conserved*hypothetical*protein Uncharacterized "ORF,*Uncharacterized"
"YOR223W,*SPAC20H4.02" Putative*protein*of*unknown*function N UHOR_04235* 4EF45 sr13695* 4EF30
pf04996 um10340* 20EF138 um10340 conserved*hypothetical*protein Uncharacterized "ORF,*Uncharacterized"
"YHR097C,*SPBC6B1.03c" Putative*protein*of*unknown*function;*green*fluorescent*protein*(GFP)Ffusion*protein*localizes*to*the*cytoplasm*and*the*nucleus
N UHOR_04674* 50EF147 sr14034* 200EF150
pf05071 um10343* 000E+0 um10343 conserved*hypothetical*protein Uncharacterized LEE1 "YPL054W,*SPAC3A11.02" ZincFfinger*protein*of*unknown*function N UHOR_04699* 50EF126 sr14051* 000E+0pf04775 um11238* 60EF48 um11238 conserved*hypothetical*protein Uncharacterized "ORF,*
Verified""YHR087W,*SPBC21C3.19" "Protein*of*unknown*function*involved*in*RNA*metabolism;*this*single*domain*protein*has*structural*
similarity*to*the*NFterminal*domain*of*SBDS,*the*human*protein*mutated*in*ShwachmanFDiamond*Syndrome*(the*yeast*SBDS*ortholog*=*SDO1/YLR022C)"
N UHOR_08954* 100EF45 sr16852* 8EF48
pf06372 um11403* 400EF165 um11403 conserved*hypothetical*protein Uncharacterized MSC1 YML128C "Protein*of*unknown*function;*mutant*is*defective*in*directing*meiotic*recombination*events*to*homologous*chromatids;*the*authentic,*nonFtagged*protein*is*detected*in*highly*purified*mitochondria*and*is*phosphorylated"
N UHOR_03756* 7EF168 sr13484* 90EF177
pf05796 um11685* 700EF69 um11685 conserved*hypothetical*protein Uncharacterized LSM12 "YHR121W,*SPBC9B6.12C*" Protein*of*unknown*function*that*may*function*in*RNA*processing;*interacts*with*Pbp1p*and*Pbp4p*and*associates*with*ribosomes;*contains*an*RNAFbinding*LSM*domain*and*an*AD*domain;*GFPFfusion*protein*is*induced*by*the*DNAFdamaging*agent*MMS
N UHOR_02350* 40EF69 sr12654* 600EF69
-
Supplemental*Dataset*4.*Gene*counts*for*CAZy*families.Family' P.#flocculosa U.#maydis U.#hordei S.#reilianum Group ActivitiesGH33 1 0 0 0 sialidase*or*neuraminidase*(EC*3.2.1.18);*transDsialidase*(EC*2.4.1.D);*2DketoD3Ddeoxynononic*acid*sialidase*(EC*3.2.1.D)GH106 1 0 0 0 alphaDLDrhamnosidase*(EC*3.2.1.40)GH113 1 0 0 0 betaDmannanase*(EC*3.2.1.78)GT77 1 0 0 0 alphaDxylosyltransferase*(EC*2.4.2.39);*alphaD1,3Dgalactosyltransferase*(EC*2.4.1.37);*arabinosyltransferase*(EC*2.4.2.D)CE2 1 0 0 0 Hemicellulose acetyl*xylan*esterase*(EC*3.1.1.72).PL4 1 0 0 0 Pectin rhamnogalacturonan*lyase*(EC*4.2.2.D).PL3 1 0 0 0 Pectin pectate*lyase*(EC*4.2.2.2).GH78 1 0 0 0 Pectin alphaDLDrhamnosidase*(EC*3.2.1.40)CBM16 1 0 0 0 CarbohydrateDbinding*module*16.*Binding*to*cellulose*and*glucomannan*demonstrated*[B.*Bae*et*al*(2008)*J*Biol*Chem.*CE12 1 0 0 0 Hemicellulose*or*pectin*side*chains pectin*acetylesterase*(EC*3.1.1.D);*rhamnogalacturonan*acetylesterase*(EC*3.1.1.D);*acetyl*xylan*esterase*(EC*3.1.1.72)CE16 2 0 0 0 Hemicellulose acetylesterase*(EC*3.1.1.6)*active*on*various*carbohydrate*acetyl*estersGH53 1 0 0 0 Hemicellulose*or*pectin*side*chains endoDbetaD1,4Dgalactanase*(EC*3.2.1.89).GH127 1 0 0 0 No*descriptionCBM5 1 0 0 0 Modules*of*approx.*60*residues*found*in*bacterial*enzymes.*ChitinDbinding*described*in*several*cases.*Distantly*related*to*the*GH28 5 1 1 2 Pectin polygalacturonase*(EC*3.2.1.15);*exoDpolygalacturonase*(EC*3.2.1.67);*exoDpolygalacturonosidase*(EC*3.2.1.82);*GH1 1 0 0 1 P/FCWDE betaDglucosidase*(EC*3.2.1.21);*betaDgalactosidase*(EC*3.2.1.23);*betaDmannosidase*(EC*3.2.1.25);*betaDglucuronidase*(EC*
3.2.1.31);*betaDDDfucosidase*(EC*3.2.1.38);*phlorizin*hydrolase*(EC*3.2.1.62);*exoDbetaD1,4Dglucanase*(EC*3.2.1.74);*6DphosphoDbetaDgalactosidase*(EC*3.2.1.85);*6DphosphoDbetaDglucosidase*(EC*3.2.1.86);*strictosidine*betaDglucosidase*(EC*3.2.1.105);*
GH93 1 0 1 0 Hemicellulose*or*pectin*side*chains exoDalphaDLD1,5Darabinanase*(EC*3.2.1.D)CBM18 3 1 1 1 Modules*of*approx.*40*residues.*The*chitinDbinding*function*has*been*demonstrated*in*many*cases.*These*modules*are*found*CBM35 2 1 0 1 Modules*of*approx.*130*residues.*A*module*that*is*conserved*in*three*Cellvibrio*xylanDdegrading*enzymes*binds*to*xylan*and*CBM50 4 2 2 1 Modules*of*approx.*50*residues*found*attached*to*various*enzymes*from*families*GH18,*GH19,*GH23,*GH24,*GH25*and*GH73,*
i.e.*enzymes*cleaving*either*chitin*or*peptidoglycan.*Binding*to*chitopentaose*demonstrated*in*the*case*of*Pteris*ryukyuensis*GH79 2 1 1 1 betaDglucuronidase*(EC*3.2.1.31);*betaD4DODmethylDglucuronidase*(EC*3.2.1.D);*heparanase*(EC*3.2.1.D);*baicalin*betaDGH43 6 3 2 4 Hemicellulose*or*pectin*side*chains betaDxylosidase*(EC*3.2.1.37);*betaD1,3Dxylosidase*(EC*3.2.1.D);*alphaDLDarabinofuranosidase*(EC*3.2.1.55);*arabinanase*(EC*GT28 2 1 1 1 1,2Ddiacylglycerol*3DbetaDgalactosyltransferase*(EC*2.4.1.46);*1,2Ddiacylglycerol*3DbetaDglucosyltransferase*(EC*2.4.1.157);**GH35 3 1 2 2 Hemicellulose betaDgalactosidase*(EC*3.2.1.23);*exoDbetaDglucosaminidase*(EC*3.2.1.165)GH18 7 4 4 4 FCWDE chitinase*(EC*3.2.1.14);*endoDbetaDNDacetylglucosaminidase*(EC*3.2.1.96);*xylanase*inhibitor;*concanavalin*B;*GH31 5 3 3 3 Hemicellulose alphaDglucosidase*(EC*3.2.1.20);*alphaD1,3Dglucosidase*(EC*3.2.1.84);*sucraseDisomaltase*(EC*3.2.1.48)*(EC*3.2.1.10);*alphaDGH2 2 1 2 1 P/FCWDE betaDgalactosidase*(EC*3.2.1.23)*;*betaDmannosidase*(EC*3.2.1.25);*betaDglucuronidase*(EC*3.2.1.31);*mannosylglycoprotein*GH117 1 0 1 1 alphaD1,3DLDneoagarooligosaccharide*hydrolase*(EC*3.2.1.D);*alphaD1,3DLDneoagarobiase*/*neoagarobiose*hydrolase*(EC*3.2.1.D)GH125 1 1 0 1 exoDalphaD1,6Dmannosidase*(EC*3.2.1.D)CE5 4 4 2 3 Hemicellulose acetyl*xylan*esterase*(EC*3.1.1.72);*cutinase*(EC*3.1.1.74)GH5 18 14 13 14 P/FCWDE chitosanase*(EC*3.2.1.132);*betaDmannosidase*(EC*3.2.1.25);*Cellulase*(EC*3.2.1.4);*glucan*1,3DbetaDglucosidase*(EC*3.2.1.58);*
licheninase*(EC*3.2.1.73);*glucan*endoD1,6DbetaDglucosidase*(EC*3.2.1.75);*mannan*endoDbetaD1,4Dmannosidase*(EC*3.2.1.78);*GH32 3 2 3 2 invertase*(EC*3.2.1.26);*endoDinulinase*(EC*3.2.1.7);*betaD2,6Dfructan*6Dlevanbiohydrolase*(EC*3.2.1.64);*endoD
levanase*(EC*3.2.1.65);*exoDinulinase*(EC*3.2.1.80);*fructan*betaD(2,1)Dfructosidase/1Dexohydrolase*(EC*3.2.1.153);*fructan*betaD(2,6)Dfructosidase/6Dexohydrolase*(EC*3.2.1.154);*sucrose:sucrose*1Dfructosyltransferase*(EC*2.4.1.99);*GT1 3 3 2 2 UDPDglucuronosyltransferase*(EC*2.4.1.17);*2Dhydroxyacylsphingosine*1DbetaDgalactosyltransferase*(EC*2.4.1.45);*NDacylsphingosine*galactosyltransferase*(EC*2.4.1.47);*flavonol*3DODglucosyltransferase*(EC*2.4.1.91);*indoleD3Dacetate*betaDglucosyltransferase*(EC*2.4.1.121);*sterol*glucosyltransferase*(EC*2.4.1.173);*ecdysteroid*UDPDglucosyltransferase*(EC*2.4.1.D);*zeaxanthin*glucosyltransferase*(EC*2.4.1.D);*zeatin*ODbetaDglucosyltransferase*(EC*2.4.1.203);*zeatin*ODbetaDxylosyltransferase*(EC*2.4.2.40);*limonoid*glucosyltransferase*(EC*2.4.1.210);*sinapate*1DGT4 5 4 4 4 sucrose*synthase*(EC*2.4.1.13);*sucroseDphosphate*synthase*(EC*2.4.1.14);*alphaDglucosyltransferase*(EC*2.4.1.52);*lipopolysaccharide*NDacetylglucosaminyltransferase*(EC*2.4.1.56);*GDPDMan*alphaDmannosyltransferase*(EC*2.4.1.D);*1,2Ddiacylglycerol*3Dglucosyltransferase*(EC*2.4.1.157);**diglucosyl*diacylglycerol*synthase*(EC*2.4.1.208);*GT15 2 2 1 2 glycolipid*2DalphaDmannosyltransferase*(EC*2.4.1.131);*GDPDMan:*alphaD1,2Dmannosyltransferase*(EC*2.4.1.D).
GH109 6 6 3 6 alphaDNDacetylgalactosaminidase*(EC*3.2.1.49)GH17 2 2 1 2 FCWDE glucan*endoD1,3DbetaDglucosidase*(EC*3.2.1.39);*glucan*1,3DbetaDglucosidase*(EC*3.2.1.58);*licheninase*(EC*3.2.1.73);*GH128 6 5 5 5 No*description
-
CBM4 2 1 2 2 Modules*of*approx.*150*residues*found*in*bacterial*enzymes.*Binding*of*these*modules*has*been*demonstrated*
CE4 9 8 6 9 FCWDE acetyl*xylan*esterase*(EC*3.1.1.72);**chitin*deacetylase*(EC*3.5.1.41);*chitooligosaccharide*deacetylase*(EC*3.5.1.D);*
GH30 3 3 2 3 glucosylceramidase*(EC*3.2.1.45);*betaD1,6Dglucanase*(EC*3.2.1.75);*betaDxylosidase*(EC*3.2.1.37);*betaDfucosidase*
GH51 3 2 3 3 Hemicellulose*or*pectin*side*chains alphaDLDarabinofuranosidase*(EC*3.2.1.55);*endoglucanase*(EC*3.2.1.4)
GH3 4 3 4 4 P/FCWDE betaDglucosidase*(EC*3.2.1.21);*xylan*1,4DbetaDxylosidase*(EC*3.2.1.37);*betaDNDacetylhexosaminidase*(EC*3.2.1.52);*
GH13 4 4 3 4 alphaDamylase*(EC*3.2.1.1);*pullulanase*(EC*3.2.1.41);*cyclomaltodextrin*glucanotransferase*(EC*2.4.1.19);*
cyclomaltodextrinase*(EC*3.2.1.54);*trehaloseD6Dphosphate*hydrolase*(EC*3.2.1.93);*oligoDalphaDglucosidase*(EC*
3.2.1.10);*maltogenic*amylase*(EC*3.2.1.133);*neopullulanase*(EC*3.2.1.135);*alphaDglucosidase*(EC*3.2.1.20);*
maltotetraoseDforming*alphaDamylase*(EC*3.2.1.60);*isoamylase*(EC*3.2.1.68);*glucodextranase*(EC*3.2.1.70);*GH16 18 21 15 16 FCWDE xyloglucan:xyloglucosyltransferase*(EC*2.4.1.207);*keratanDsulfate*endoD1,4DbetaDgalactosidase*(EC*3.2.1.103);*endoD
GH39 2 1 3 2 Hemicellulose alphaDLDiduronidase*(EC*3.2.1.76);*betaDxylosidase*(EC*3.2.1.37).
GH37 3 3 3 3 alpha,alphaDtrehalase*(EC*3.2.1.28).
GH20 2 2 2 2 FCWDE betaDhexosaminidase*(EC*3.2.1.52);*lactoDNDbiosidase*(EC*3.2.1.140);*betaD1,6DNDacetylglucosaminidase)*(EC*3.2.1.D
GH27 1 1 0 2 Hemicellulose alphaDgalactosidase*(EC*3.2.1.22);*alphaDNDacetylgalactosaminidase*(EC*3.2.1.49);*isomaltoDdextranase*(EC*
CBM63 1 1 1 1 No*description
GH8 1 1 1 1 chitosanase*(EC*3.2.1.132);*cellulase*(EC*3.2.1.4);*licheninase*(EC*3.2.1.73);*endoD1,4DbetaDxylanase*(EC*3.2.1.8);*
GH115 1 1 1 1 Pectin xylan*alphaD1,2Dglucuronidase*(3.2.1.131);*alphaD(4DODmethyl)Dglucuronidase*(3.2.1.D)
GH15 1 1 1 1 glucoamylase*(EC*3.2.1.3);*glucodextranase*(EC*3.2.1.70);*alpha,alphaDtrehalase*(EC*3.2.1.28)
GT76 1 1 1 1 DolDPDMan:*alphaD1,6Dmannosyltransferase*(EC*2.4.1.D)
PL12 1 1 1 1 heparinDsulfate*lyase*(EC*4.2.2.8)
CE9 1 1 1 1 NDacetylglucosamine*6Dphosphate*deacetylase*(EC*3.5.1.25);*NDacetylgalactosamineD6Dphosphate*deacetylase*(EC*
GT66 1 1 1 1 DolDPPDalphaDoligosaccharide:*protein*betaDoligosaccharyltransferase*(EC*2.4.1.119)
GH92 3 3 3 3 FCWDE mannosylDoligosaccharide*alphaD1,2Dmannosidase*(EC*3.2.1.113);*mannosylDoligosaccharide*alphaD1,3Dmannosidase*
PL1 1 1 1 1 Pectin pectate*lyase*(EC*4.2.2.2);*exoDpectate*lyase*(EC*4.2.2.9);*pectin*lyase*(EC*4.2.2.10).
GH74 1 1 1 1 Cellulose endoglucanase*(EC*3.2.1.4);*oligoxyloglucan*reducing*endDspecific*cellobiohydrolase*(EC*3.2.1.150);*xyloglucanase*
GT57 2 2 2 2 DolDPDGlc:*alphaD1,3Dglucosyltransferase*(EC*2.4.1.D)
GH72 1 1 1 1 FCWDE betaD1,3Dglucanosyltransglycosylase*(EC*2.4.1.D)
GT58 1 1 1 1 DolDPDMan:*dolichol*pyrophosphateDmannose*alphaD1,3Dmannosyltransferase*(EC*2.4.1.130);*DolDPDMan:*dolichol*
GT59 1 1 1 1 DolDPDGlc:*Glc2Man9GlcNAc2DPPDDol*alphaD1,2Dglucosyltransferase*(EC*2.4.1.D)
GT3 1 1 1 1 glycogen*synthase*(EC*2.4.1.11).
GT8 3 3 3 3 lipopolysaccharide*alphaD1,3Dgalactosyltransferase*(EC*2.4.1.44);**UDPDGlc:*(glucosyl)lipopolysaccharide*alphaD1,2D
glucosyltransferase*(EC*2.4.1.D);*lipopolysaccharide*glucosyltransferase*1*(EC*2.4.1.58);*glycogenin*
glucosyltransferase*(EC*2.4.1.186);*inositol*1DalphaDgalactosyltransferase*(galactinol*synthase)*(EC*2.4.1.123);*GH85 1 1 1 1 FCWDE endoDbetaDNDacetylglucosaminidase*(EC*3.2.1.96)
GT50 1 1 1 1 DolDPDMan*alphaD1,4Dmannosyltransferase*(EC*2.4.1.D)
GH62 1 1 1 1 Hemicellulose*or*pectin*side*chains alphaDLDarabinofuranosidase*(EC*3.2.1.55)
CBM21 1 1 1 1 Modules*of*approx.*100*residues.*The*granular*starchDbinding*function*has*been*demonstrated*in*one*case.*
GT48 1 1 1 1 1,3DbetaDglucan*synthase*(EC*2.4.1.34)
CE14 1 1 1 1 NDacetylD1DDDmyoDinositylD2DaminoD2DdeoxyDalphaDDDglucopyranoside*deacetylase*(EC*3.5.1.89);*diacetylchitobiose*
CBM33 1 1 1 1 Binding*to*chitin*has*been*demonstrated*in*several*cases*and*binding*to*chitosan*descibed*in*a*single*case.
GH55 1 1 1 1 FCWDE exoDbetaD1,3Dglucanase*(EC*3.2.1.58);*endoDbetaD1,3Dglucanase*(EC*3.2.1.39).
GT32 2 2 2 2 alphaD1,6Dmannosyltransferase*(EC*2.4.1.D);*alphaD1,4DNDacetylglucosaminyltransferase*(EC*2.4.1.D);*alphaD1,4DND
GT33 1 1 1 1 GDPDMan:*chitobiosyldiphosphodolichol*betaDmannosyltransferase*(EC*2.4.1.142).
GT39 3 3 3 3 DolDPDMan:*protein*alphaDmannosyltransferase*(EC*2.4.1.109)
GH47 3 3 3 3 alphaDmannosidase*(EC*3.2.1.113)
GH42 1 1 1 1 betaDgalactosidase*(EC*3.2.1.23)
GT24 1 1 1 1 UDPDGlc:*glycoprotein*alphaDglucosyltransferase*(EC*2.4.1.D).
GT22 4 4 4 4 DolDPDMan*alphaDmannosyltransferase*(EC*2.4.1.D)
CBM43 1 1 1 1 Modules*of*approx.*90D100*residues*found*at*the*CDterminus*of*GH17*or*GH72*enzymatic*modules*and*also*
GT20 3 3 3 3 alpha,alphaDtrehaloseDphosphate*synthase*[UDPDforming]*(EC*2.4.1.15)
CBM48 1 1 1 1 Modules*of*approx.*100*residues*with*glycogenDbinding*function,*appended*to*GH13*modules.*Also*found*in*the*
-
GT45 2 2 2 2 alphaDNDacteylglucosaminyltransferase*(EC*2.4.1.D)
CE10 27 31 23 28 arylesterase*(EC*3.1.1.D);**carboxyl*esterase*(EC*3.1.1.3);*acetylcholinesterase*(EC*3.1.1.7);*cholinesterase*(EC*
CE1 19 22 18 22 Hemicellulose acetyl*xylan*esterase*(EC*3.1.1.72);*cinnamoyl*esterase*(EC*3.1.1.D);*feruloyl*esterase*(EC*3.1.1.73);*carboxylesterase*
GT2 9 10 10 10 cellulose*synthase*(EC*2.4.1.12);*chitin*synthase*(EC*2.4.1.16);*dolichylDphosphate*betaDDDmannosyltransferase*(EC*
2.4.1.83);*dolichylD