Supplemental Data. Lei$etal.$(2013).$PlantCell$10.1105/tpc ... · 12/23/2013 · Supplemental...
Transcript of Supplemental Data. Lei$etal.$(2013).$PlantCell$10.1105/tpc ... · 12/23/2013 · Supplemental...

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
Supplemental Figure 1. CSI1-like proteins in Arabidopsis. (A) Structure of CSI1 and CSI1-like proteins. Orange bar represents the Armadillo (ARM) repeat and green rectangle represents the C2 domain. (B) Expression profile of CSI1, CSI2, and CSI3. Data were obtained from Genevestigator (www.genevestigator..com). Signal intensity of Y-axis represents arbitrary units. From the left to the right, black bar represents expression in tissues including callus (1), cell suspension (2), seedling (3), cotyledons (4), hypocotyl (5), radicle (6), inflorescene (7), flower (8), carpel (9), ovary (10), stigma (11), petal (12), sepal (13), stamen (14), pollen (15), pedicle (16), silique (17), seed (18), embryo (19), stem (20), node (21), shoot apex (22), cauline leaf (23), rosette (24), juvenile leaf (25), adult leaf (26), petiole (27), senescent leaf (28), xylem (29), cork (30), rots (31), primary root (32), lateral root (33), root tip (34), elongation zone (35), root hair zone (36), endodermis (37), endodermis + cortex (38), epidermal atrichoblasts (39), lateral root cap (40), and stele (41).

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
2
Supplemental Figure 2. CSI3 is expressed in various tissues. The ProCSI3:GUS construct was transformed into Arabidopsis (Col-0) plants. GUS expression was detected by histochemical staining. (A) Light-grown 6-day-old seedlings. Scale bar = 1 mm. Inset shows GUS staining at the root tip. Scale bar = 200 µm. (B) Four-day-old dark-grown hypocotyls. Scale bar = 200 µm. (C) Four-week-old mature rosette leaf. Scale bar = 1 mm. Inset shows GUS staining in trichomes. Scale bar = 200 µm. (D) Soil-grown roots from 6-week-old plants. Scale bar = 2 mm. Inset shows GUS staining in the root tip. Scale bar = 200 µm. (E and F) Mature flowers before and after pollination, respectively. Scale bar = 200 µm. Inset shows GUS staining in anthers. Scale bar = 200 µm. (G) Siliques. Scale bar = 1 mm.

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
3
Supplemental Figure 3. T-DNA insertion analysis of csi3-1 mutant. (A) Schematic representation of T-DNA insertion site in csi3-1 mutant. (B) RT-PCR analysis of mRNA expression in csi3 mutants. ACTIN2 (ACT2) was used as control. WT, Columbia-0. The result showed is a representative image from three biological replicates.
Supplemental Figure 4. Anisotropic growth defect and cellulose content of the csi1 csi3 double mutant. (A) Quantification of hypocotyl width of 4-day-old dark-grown wild type (Col-0), csi1, csi3, and csi1 csi3. Data are based on ~50 seedlings for each genotype. (B) Crystalline cellulose content of 4-day-old dark-grown wild type (Col-0), csi1, csi3, and csi1 csi3. Error bars represent standard deviation (SD). Double asterisks indicate a significant difference from WT at the same data point (p < 0.01; Student’s t-test).

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
4
Supplemental Figure 5. Similar to CSI1, CSI3 is localized to distinct particles at the plasma membrane. (A) Schematic representation of ProCSI3:GFP-CSI3. (B) Four-day-old dark-grown seedlings of Col-0, csi1, csi1 csi3, ProCSI3:GFP-CSI3 in csi1 line #4 (4), and ProCSI3:GFP-CSI3 in csi1 line #11 (11). Scale bar = 5 mm. (C) Quantification of hypocotyl length of dark-grown seedlings shown in (B). Data are based on ~50 seedlings for each genotype. (D) Scanning electron micrograph of four-day-old, dark-grown hypocotyls from Col-0, csi1, csi1 csi3, ProCSI3:GFP-CSI3 in csi1 line #4 , and ProCSI3:GFP-CSI3 in csi1 line #11 (from left to right). Scale bar =100 µm. (E) Optical sections of epidermal cells in 3-day-old, dark-grown hypocotyls expressing GFP-CSI1 or GFP-CSI3. Focal planes of the plasma membrane and lower cortex are shown. Scale bar = 5µm.

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
5
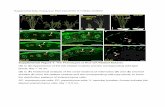
Supplemental Figure 6. CSI3 partially associates with SmaCCs/MASCs. Displayed are two-channel imaging of epidermal cells co-expressing RFP-CSI1 and GFP-CSI3 upon 100 nM isoxaben treatment for 2 h. (A) Single optical section of RFP-CSI1, GFP-CSI3, and merged image, and colocalization analysis. A representative image from 47 cells from 17 seedlings is shown. White dots (n = 19) represent co-localized RFP-CSI1 and GFP-CSI3. RFP-CSI1 or GFP-CSI3 that did not colocalize is in red (n = 24) and green (n = 44), respectively. Scale bar = 5 µm. (B) Plot of a line scan showing spatial correlation between RFP-CSI1 and GFP-CSI3 signals. The line is indicated in the merged image in (A). (C) Dynamic behavior of SmaCCs/MASCs. Note that SmaCCs/MASCs labeled with RFP-CSI1 and GFP-CSI3 underwent merging and splitting events. Scale bar = 0.5 µm. (D) Histogram of SmaCCs/MASCs velocities.

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
6
Supplemental Figure 7. Co-localization analyses among CSI1, CSI3, and microtubules. (A) Two-channel imaging of Arabidopsis seedlings co-expressing GFP-CSI3 and RFP-TUA5 and quantification of colocalization. A representative image from 71 cells from 37 seedlings is shown. White dots (n = 131) represent co-localized GFP-CSI3 with microtubules. GFP-CSI3 that did not colocalize with microtubules are colored in blue (n = 27). Scale bar = 5 µm. (B) Three-channel imaging of Arabidopsis seedlings co-expressing CFP-TUA1, GFP-CSI3, and RFP-CSI1. Shown is a representative image from three cells from three seedlings. In the colocalization analysis of a representative merged image, white dots (n = 51) represent colocalized CFP-TUA1, GFP-CSI3, and RFP-CSI1. Pink dots (n = 120) represent colocalized RFP-CSI1 and CFP-TUA1, but not GFP-CSI3. Light blue dots (n = 93) represent colocalized GFP-CSI3 and CFP-TUA1, but not RFP-CSI1. Yellow dots (n = 68) represent colocalized RFP-CSI1 and GFP-CSI3, but not CFP-TUA1. Red dots (n = 25) represent RFP-CSI1 that did not colocalize with other signals. Dark blue (n = 32) dots represent GFP-CSI3 that did not colocalize with other signals. Scale bar = 5 µm.

Supplemental Data. Lei et al. (2013). Plant Cell 10.1105/tpc.113.116715
7
Supplemental Table 1. DNA primers used in this study. T-DNA lines Primer sequence (5’-3’) csi1-3 (SALK_138584)
F: TAGGCACACCAAAAGATGAAC R: GAGCTACAGAGCCTGCAACAC
csi2-1 (FLAG_209F11)
F: ATGACGTCTGAGATGGATGATCCAGAG R: GTTCGCTAGACAAACCAAGGTACG
csi3-1 (GABI_308G07)
F: GTAAATCTATCCCGCTGCTTGTAG R: AGCTCGTCTATTCGTTCTACTATTCATCAC
SWAP constructs ProCSI1 F: GAGCTCAATTTTTGAATTTCGCCAAAACGTTTACC
R: ACTAGTATCTTCACTTCACTTAAAAAATTCTCAAATC ProCSI1:GFP-CSI3 F: CGGCAAACTAAGGTCGTGAGC
R: AAGTCGTGCTGCTTCATGTG FP-fusion constructs ProCSI1:CSI1-GFP F: GAGCTCAATTTTTGAATTTCGCCAAAACGTTTACC
R: ACTAGTATCTTCACTTCACTTAAAAAATTCTCAAATC ProCSI3:GFP-CSI3 F: GAGCTCGAGATCCGATTTTACTGACTG
R: CGGATATCCGCTTTTAGCTTCAGCCTCTGG ProCSI3:CSI3-RFP F: GAGCTCGAGATCCGATTTTACTGACTG
R: CGGATATCCGCTTTTAGCTTCAGCCTCTGG Y2H constructs CSI1 F: ATGACAAGTGCTCTTGGA
R: CTTGTTAGACCACTGGAATT CSI3 F: ATGGAAATGGACGACCCA
R: TCAATGAGTCTCATCCGTTG CESA3 F: GAAATGGAATCCGAAGGGGAAA
R: GGAACAGTTGATTCCACATTC CESA6 F: GCAATGAACACCGGTGGTCGGT
R: GGACAAGCAGTCTAAACCACA GUS constructs ProCSI2 F: GAGCTCCTAGTCGTGGCTAAGTTAGAAGGTC
R: CGACTAGTTGTTTTATTATCGTCCCTGCACCC ProCSI3 F: GAGCTCGAGATCCGATTTTACTGACTG
R: CGGATATCCGCTTTTAGCTTCAGCCTCTGG RT-PCR ACTIN2 F: CTAGGATCCAAAATGGCCGATGGTGAGG
R: GAGACTCACCACCACGAACCAG CSI3 F: GGTCATTGTTATGGAAGCTGGC
R: GGTCTTCTCGCGATTACTCCACTATTTGTT