Supplemental Figures - Biochemistry · Supplemental Figures: Figure S1. Analysis of endo-siRNA...
Transcript of Supplemental Figures - Biochemistry · Supplemental Figures: Figure S1. Analysis of endo-siRNA...

Supplemental Figures:
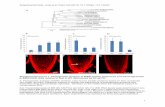
Figure S1. Analysis of endo-siRNA targets in different microarray datasets. The
percentage of each array dataset that were predicted endo-siRNA targets according to the
Ambros dataset (Lee et al. 2006) was plotted [(number of endo-siRNA targets in
microarray dataset / total genes in microarray dataset) * 100]. Data are also presented for
the datasets after parsing into upregulated and downregulated genes. Additionally, the
genes common to two or more of the lists of upregulated genes were also analyzed (∩
indicates intersection between datasets). The random analysis represents the average
value using ten independently generated random lists of genes (see Experimental
Procedures).

Figure S2. Chromosomal location and strand orientation of all misregulated genes in the
dcr-1(-/-) dataset. Horizontal black line depicts chromosome. Vertical bars indicate
misregulated genes, depicted above and below the chromosome to denote opposite strand
orientations.

Figure S3. Figure of significant GO terms that are under-represented among
misregulated genes in animals that lack dcr-1, rde-4 or rde-1. See legend to Figure 3 for
additional details.

Supplemental Table Legends
Table S1
Datasets of genes determined to be misregulated by microarray analyses of dcr-1(-/-),
rde-4(-/-), and rde-1(-/-) mutants. A separate worksheet was created for each curated list
of genes and shows the Affymetrix probeset ID, the fold change, and direction of
misregulation. A separate worksheet was also included for analyses of the intersections
between the three datasets. In addition, for each microarray analysis the complete output
from the microarray analysis suite (GeneSifter) is provided as a separate worksheet.
Table S2
Analyses of PicTar predicted miRNA targets in the dcr-1(-/-), rde-4(-/-), and rde-1(-/-)
datasets. The accumulated list of PicTar predicted miRNA targets (cePicTar) that was
generated from data obtained from the PicTar website (http://pictar.bio.nyu.edu/cgi-
bin/new_PicTar_nematode.cgi?species=nematode) is given in column A of the worksheet
“cePicTar”. A description of how the cePicTar list was generated, as well as the raw
numbers from the miRNA analysis are presented in the worksheet “methods.” Column A
of worksheet “PicTar AND dcr upregulated” lists the subset of all PicTar predicted
miRNA targets that are upregulated in the dcr-1(-/-) dataset. The PicTar predicted let-7
targets are listed in column A of the worksheet “PicTar let-7” with the dcr-1(-/-)
upregulated genes highlighted in blue.
Table S3
Analysis of the overlap between previously published infection study datasets (Huffman
et al. 2004; O'Rourke et al. 2006; Shapira et al. 2006) and dcr-1(-/-), rde-4(-/-), and rde-
1(-/-) datasets. The lists of genes used for each infection study microarray are presented

in worksheet “immunity lists” with the upregulated and downregulated genes denoted in
red or green, respectively. The quantitative analysis of overlap between the microarray
datasets are presented in the worksheet “intersection of microarrays.” Shown in Section
A are the number of misregulated genes that are common to both of the datasets being
compared. The number of genes expected to be common as a result of random chance is
shown in Section B. The ratio of observed (Section A) / expected (Section B) is presented
in Section C. Additionally, the genes used to determine misregulated members of families
linked to innate immunity are listed in worksheet “innate immunity families” (columns
N-AH). The number of family members misregulated in each of the datasets analyzed
(columns B-J) is presented along with the details of the analysis.
Table S4
This dataset shows the lists submitted to Fatigo+ for analysis. For each mutant animal
analyzed, column A (init_genes 1) corresponds to the genes misregulated ≥ 1.5 fold and
column B (init_genes 2) corresponds to the remainder of genes on the array.
Table S5
This dataset includes the complete Fatigo+ analysis for each mutant animal. Values
reported in columns C and F indicate the number of genes found with the specified GO
term among the ≥ 1.5 fold misregulated genes (column C) or the remainder of the genes
on the array (column F, total genes on the array excluding those on the ≥ 1.5 fold list).
Values in columns C and F were divided by the total number of genes with annotation at
the specified ontological level to determine the percentage values shown in columns D
and G. For the percentage values plotted in Figures 3 and S3, the total genes at the
specified ontological level are as follows. dcr-1, ≥1.5 fold: level 3, 437 (mf), 465(bp) and

231(cc), and level 4, 377(mf), 432(bp) and 159(cc); array minus ≥ 1.5 fold: level 3,
5219(mf), 5346(bp) and 2906(cc), and level 4, 4432(mf), 5079(bp), and 1976(cc). rde-4,
≥1.5 fold: level 3, 140(mf), 130(bp), 42(cc), and level 4, 119(mf), 109(bp) and 28(cc);
array minus ≥1.5 fold: level 3, 5515(mf), 5678(bp), 3090(cc); level 4, 4684(mf),
5397(bp) and 2108(cc). rde-1, ≥1.5 fold: level 3, 25(mf), 25(bp), 10(cc) and level 4,
23(mf), 25(bp), 8(cc); array minus ≥ 1.5 fold: level 3, 5629(mf), 5782(bp) and 3127(cc)
and level 4, 4780 (mf), 5482 (bp), 2128 (cc).