Computational Biology, Part 3 Sequence Alignment Robert F. Murphy Copyright 1996, 1999-2009. All...
-
Upload
stella-todd -
Category
Documents
-
view
214 -
download
0
Transcript of Computational Biology, Part 3 Sequence Alignment Robert F. Murphy Copyright 1996, 1999-2009. All...
Computational Biology, Part 3Sequence Alignment
Computational Biology, Part 3Sequence Alignment
Robert F. MurphyRobert F. Murphy
Copyright Copyright 1996, 1999-2009. 1996, 1999-2009.
All rights reserved.All rights reserved.
Sequence AlignmentSequence Alignment
Definition: Procedure for comparing two or Definition: Procedure for comparing two or more sequences by searching for a series of more sequences by searching for a series of individual characters or character patterns individual characters or character patterns that are that are in the same orderin the same order in the sequences in the sequences Pair-wise alignmentPair-wise alignment: compare two sequences: compare two sequences Multiple sequence alignmentMultiple sequence alignment: compare more : compare more
than two sequencesthan two sequences
Example sequence alignmentExample sequence alignment
Task: align Task: align “abcdef”“abcdef” with with “abdgf”“abdgf” Write second sequence below the firstWrite second sequence below the first
abcdefabcdefabdgfabdgf
Move sequences to give maximum match between Move sequences to give maximum match between themthem
Show characters that match using vertical barShow characters that match using vertical bar
Example sequence alignmentExample sequence alignment
abcdefabcdef
||||
abdgfabdgf Insert gap between Insert gap between bb and and dd on lower on lower
sequence to allow sequence to allow dd and and ff to align to align
Example sequence alignmentExample sequence alignment
abcdefabcdef
|| | ||| | |
ab-dgfab-dgf Note Note ee and and gg don’t match don’t match
Matching Similarity vs. IdentityMatching Similarity vs. Identity
Alignments can be based on finding only Alignments can be based on finding only identical characters, or (more commonly) identical characters, or (more commonly) can be based on finding can be based on finding similarsimilar characters characters
More on how to define More on how to define similaritysimilarity later later
Global vs. Local AlignmentGlobal vs. Local Alignment
We distinguishWe distinguish GlobalGlobal alignment algorithms which optimize alignment algorithms which optimize
overall overall alignment between two sequences alignment between two sequences LocalLocal alignment algorithms which seek only alignment algorithms which seek only
relatively relatively conservedconserved pieces of sequence pieces of sequence Alignment stops at the ends of regions of strong Alignment stops at the ends of regions of strong
similaritysimilarity Favors finding conserved patterns in otherwise Favors finding conserved patterns in otherwise
different pairs of sequencesdifferent pairs of sequences
Global vs. Local AlignmentGlobal vs. Local Alignment
GlobalGlobal
LGPSSKQTGKGS-SRIWDNLGPSSKQTGKGS-SRIWDN| | ||| | | | | ||| | | LN-ITKSAGKGAIMRLGDALN-ITKSAGKGAIMRLGDA
LocalLocal
--------GKG----------------GKG-------- ||| ||| --------GKG----------------GKG--------
Global vs. Local AlignmentGlobal vs. Local Alignment
GlobalGlobal
LGPSSKQTGKGS-SRIWDNLGPSSKQTGKGS-SRIWDN| | ||| | | | | ||| | | LN-ITKSAGKGAIMRLGDALN-ITKSAGKGAIMRLGDA
LocalLocal
-------TGKG---------------TGKG-------- ||| ||| -------AGKG---------------AGKG--------
Why do sequence alignments?Why do sequence alignments?
To find whether two (or more) genes or To find whether two (or more) genes or proteins are evolutionarily related to each proteins are evolutionarily related to each otherother
To find structurally or functionally similar To find structurally or functionally similar regions within proteinsregions within proteins
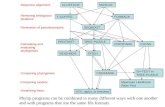
Origin of similar genesOrigin of similar genes
Similar genes arise by Similar genes arise by gene gene duplicationduplication
Copy of a gene inserted next Copy of a gene inserted next to the originalto the original
Two copies mutate Two copies mutate independentlyindependently
Each can take on separate Each can take on separate functionsfunctions
All or part can be transferred All or part can be transferred from one part of genome to from one part of genome to anotheranother
http://fig.cox.miami.edu/~cmallery/150/gene/c7.19.19.gene.family.jpg
Methods for Pairwise AlignmentMethods for Pairwise Alignment
Dot matrix analysisDot matrix analysis Dynamic ProgrammingDynamic Programming Word or Word or k-k-tuple methods (FASTA and tuple methods (FASTA and
BLAST)BLAST)
Sequence comparison with dot matricesSequence comparison with dot matrices Goal: Goal: Graphically display regions of Graphically display regions of
similarity between two sequences (e.g., similarity between two sequences (e.g., domains in common between two proteins domains in common between two proteins of suspected similar function)of suspected similar function)
Sequence comparison with dot matricesSequence comparison with dot matrices Basic Method: Basic Method: For two sequences of For two sequences of
lengths M and N, lay out an M by N grid lengths M and N, lay out an M by N grid (matrix) with one sequence across the top (matrix) with one sequence across the top and one sequence down the left side. For and one sequence down the left side. For each position in the grid, compare the each position in the grid, compare the sequence elements at the top (column) and sequence elements at the top (column) and to the left (row). If and only if they are the to the left (row). If and only if they are the same, place a dot at that position.same, place a dot at that position.
Examples for protein sequencesExamples for protein sequences (Demonstration A6, Sequence 1 vs. 2)(Demonstration A6, Sequence 1 vs. 2)
abcdaefghbijklcmnopdabcdaefghbijklcmnopd
abcdaefghbijklcmnopdabcdaefghbijklcmnopd
Interpretation of dot matricesInterpretation of dot matrices
Regions of similarity appear as diagonal Regions of similarity appear as diagonal runs of dotsruns of dots
Reverse diagonals (perpendicular to Reverse diagonals (perpendicular to diagonal) indicate inversionsdiagonal) indicate inversions
Reverse diagonals crossing diagonals (Xs) Reverse diagonals crossing diagonals (Xs) indicate palindromesindicate palindromes
Examples for protein sequencesExamples for protein sequences (Demonstration A6, Sequence 4 vs. 4)(Demonstration A6, Sequence 4 vs. 4)
abcdeedcbafghijklmno abcdeedcbafghijklmno abcdeedcbafghijklmnoabcdeedcbafghijklmno
Interpretation of dot matricesInterpretation of dot matrices
Can link or "join" separate diagonals to Can link or "join" separate diagonals to form form alignmentalignment with "gaps" with "gaps" Each a.a. or base can only be used onceEach a.a. or base can only be used once
Can't trace vertically or horizontallyCan't trace vertically or horizontally Can't double backCan't double back
A gap is introduced by each vertical or A gap is introduced by each vertical or horizontal skiphorizontal skip
Examples for protein sequencesExamples for protein sequences (Demonstration A6, Sequence 2 vs. 3)(Demonstration A6, Sequence 2 vs. 3)
abcdaefghbijklcmnopdabcdaefghbijklcmnopd
abcdefghijklmnopqrstabcdefghijklmnopqrst
Uses for dot matricesUses for dot matrices
Can use dot matrices to align two proteins Can use dot matrices to align two proteins or two nucleic acid sequencesor two nucleic acid sequences
Can use to find amino acid repeats within a Can use to find amino acid repeats within a protein by comparing a protein sequence to protein by comparing a protein sequence to itselfitself Repeats appear as a set of diagonal runs stacked Repeats appear as a set of diagonal runs stacked
vertically and/or horizontallyvertically and/or horizontally
Examples for protein sequencesExamples for protein sequences (Demonstration A6, Sequence 5 vs. 5)(Demonstration A6, Sequence 5 vs. 5)
abcdabcdabcdabcdabcdabcdabcdabcdabcdabcd
abcdabcdabcdabcdabcdabcdabcdabcdabcdabcd
Uses for dot matricesUses for dot matrices
Can use to find self base-pairing of an RNA Can use to find self base-pairing of an RNA (e.g., tRNA) by comparing a sequence to (e.g., tRNA) by comparing a sequence to itself complemented and reverseditself complemented and reversed
Excellent approach for finding sequence Excellent approach for finding sequence transpositionstranspositions
Filtering to remove “noise”Filtering to remove “noise”
A problem with dot matrices for long A problem with dot matrices for long sequences is that they can be very noisy due sequences is that they can be very noisy due to lots of insignificant matches (i.e., one A)to lots of insignificant matches (i.e., one A)
Solution use a window and a thresholdSolution use a window and a threshold compare character by character within a compare character by character within a
window (have to choose window size)window (have to choose window size) require certain fraction of matches within require certain fraction of matches within
window in order to display it with a “dot”window in order to display it with a “dot”
How do we choose a window size?How do we choose a window size? Window size changes with goal of analysisWindow size changes with goal of analysis
size of average exonsize of average exon size of average protein structural elementsize of average protein structural element size of gene promotersize of gene promoter size of enzyme active sitesize of enzyme active site
How do we choose a threshold value?How do we choose a threshold value? Threshold based on statisticsThreshold based on statistics
using shuffled actual sequenceusing shuffled actual sequence find average (find average (mm) and s.d. () and s.d. () of match scores of ) of match scores of
shuffled sequenceshuffled sequence convert original (unshuffled) scores (convert original (unshuffled) scores (xx) to) to ZZ scoresscores
• Z = (x - m)/Z = (x - m)/ use threshold Z of of 3 to 6use threshold Z of of 3 to 6
using analysis of other sets of sequencesusing analysis of other sets of sequences provides “objective” standard of significanceprovides “objective” standard of significance
Dot matrix analysis with Matlab bioinformatics toolboxDot matrix analysis with Matlab bioinformatics toolbox Get phage Get phage cI and phage P22 c2 repressor cI and phage P22 c2 repressor
sequences from Genbank (X00166 and sequences from Genbank (X00166 and V01153 respectively)V01153 respectively)
Use window size of 11 and stringency of 7Use window size of 11 and stringency of 7
Matlab codeMatlab code
getgenbank('X00166', 'TOFILE', 'HGENBANKX00166.GBK');getgenbank('X00166', 'TOFILE', 'HGENBANKX00166.GBK');
getgenbank('V01153', 'TOFILE', 'HGENBANKV01153.GBK');getgenbank('V01153', 'TOFILE', 'HGENBANKV01153.GBK');
seq1 = genbankread('HGENBANKX00166.GBK');seq1 = genbankread('HGENBANKX00166.GBK');
seq2 = genbankread('HGENBANKV01153.GBK');seq2 = genbankread('HGENBANKV01153.GBK');
window=11; num=7;window=11; num=7;
seqdotplot(seq1,seq2,window,num)seqdotplot(seq1,seq2,window,num)
xlabel('X00166');xlabel('X00166');
ylabel('V01153');ylabel('V01153');
title('Window 11 Num 7');title('Window 11 Num 7');
Dot matrixDot matrix
Note set of Note set of diagonals diagonals in lower in lower right that right that do not line do not line up due to up due to insertion insertion near 475 near 475 on cIon cI
Dot matrix analysis with DotmatcherDot matrix analysis with Dotmatcher Get the corresponding protein sequence of Get the corresponding protein sequence of
phage phage cI and phage P22 c2 repressor cI and phage P22 c2 repressor sequences (CAA24991 and CAA24470 sequences (CAA24991 and CAA24470 respectively)respectively)
Use Emboss Dotmatcher online:Use Emboss Dotmatcher online: emboss.bioinformatics.nlemboss.bioinformatics.nl
under ‘under ‘ALIGNMENT DOT PLOTS’ALIGNMENT DOT PLOTS’
Use window size of 10 and threshold of 23 Use window size of 10 and threshold of 23 BLOSUM62 units (default parameters)BLOSUM62 units (default parameters)
Dot matrixDot matrix Similarity in Similarity in
the carboxy-the carboxy-terminal terminal domains of domains of the proteins the proteins agrees with agrees with the similarity the similarity in 3’ends of in 3’ends of the two DNA the two DNA sequences.sequences.
Dot matrix analysis with Matlab bioinformatics toolboxDot matrix analysis with Matlab bioinformatics toolbox Get human LDL receptor protein sequence Get human LDL receptor protein sequence
from Genbank (P01130)from Genbank (P01130) Use window size of 1 and stringency of 1Use window size of 1 and stringency of 1 Use window size of 23 and stringency of 7Use window size of 23 and stringency of 7
Matlab codeMatlab code
getgenpept('P01130', 'TOFILE', 'HGENBANKP01130.GBK');getgenpept('P01130', 'TOFILE', 'HGENBANKP01130.GBK'); seq5 = genbankread('HGENBANKP01130.GBK');seq5 = genbankread('HGENBANKP01130.GBK'); window=1; num=1; seqdotplot(seq5,seq5,window,num)window=1; num=1; seqdotplot(seq5,seq5,window,num) xlabel('P01130 Human LDL receptor');xlabel('P01130 Human LDL receptor'); ylabel('P01130 Human LDL receptor');ylabel('P01130 Human LDL receptor'); title('Window 1 Num 1');title('Window 1 Num 1'); window=23; num=7; seqdotplot(seq5,seq5,window,num)window=23; num=7; seqdotplot(seq5,seq5,window,num) xlabel('P01130 Human LDL receptor');xlabel('P01130 Human LDL receptor'); ylabel('P01130 Human LDL receptor');ylabel('P01130 Human LDL receptor'); title('Window 23 Num 7');title('Window 23 Num 7');
Dot matrixDot matrix
W=1 S=1W=1 S=1 Note set of Note set of
stacked stacked diagonals diagonals in upper in upper leftleft