Sequence Alignment Variations
description
Transcript of Sequence Alignment Variations

Sequence Alignment Variations
• Computing alignments using only O(m) space rather than O(mn) space.
• Computing alignments with bounded difference
• Exclusion methods: fast expected running times

1. Linear Space
• Hirschberg [1977]• Suppose we only need the maximum
similarity value of S and T without an alignment or transcript
• How can we conserve space?– Only save row i-1 when computing row i in the
table

Illustration
0
1
2
3
4
n
n-1
0 1 2 3 4 5 6 7 … m
...

Linear space and an alignment• Assume S has length 2n• Divide and conquer approach
– Compute value of optimal alignment of S[1..n] with all prefixes of T
• Store row n only at end along with pointer values of row n– Compute value of optimal alignment of Sr[1..n] with all
prefixes of Tr
• Store only values in row n
• Find k such that – V(S[1..n],T[1..k]) + V(Sr[1..n],Tr[1..m-k]) – is maximized over 0 <= k <=m

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0
V(S[1..6], T[1..0])
V(Sr[1..6], Tr[1..18])
k=0
m-k=18

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0
V(S[1..6], T[1..1])
V(Sr[1..6], Tr[1..17])
k=1
m-k=17

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0
V(S[1..6], T[1..2])
V(Sr[1..6], Tr[1..16])
k=2
m-k=16

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0
V(S[1..6], T[1..9])
V(Sr[1..6], Tr[1..9])
k=9
m-k=9

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0
V(S[1..6], T[1..18])
V(Sr[1..6], Tr[1..0])
k=18
m-k=0

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0

Recursive Step
• Let k* be the k that maximizes – V(S[1..n],T[1..k]) + V(Sr[1..n],Tr[1..m-k])
• Record all steps on row n including the one from n-1 and the one to n+1
• Recurse on the two subproblems– S[1..n-1] with T[1..j] where j <= k*– Sr[1..n] with Tr[1..q] where q <= m-k*

Illustration0123456
0 1 2 3 4 5 6 7 8 9 0 1 2 3 4 5 6 7 8 -
6543210
- 8 7 6 5 4 3 2 1 0 9 8 7 6 5 4 3 2 1 0

Time Required
• cmn time to get this answer so far• Two subproblems have at most half the total
size of this problem– At most the same cmn time to get the rest of the
solution• cmn/2 + cmn/4 + cmn/8 + cmn/16 + … <= cmn/2
• Final result– Linear space with only twice as much time

Extending to local alignment
• What are the problems?• Don’t know what substrings of S and T to align, so
we won’t know midpoints• Solution
– Find end point by computing only values and storing max value (and location) along the way
– Find start point by computing a “reversed” dynamic program using the reverse strings starting at i in S and j in T
– Once end points are fixed, just like global alignment

2. Bounded Difference
• Suppose the number of differences between S and T is bounded
• Typically focus on (unweighted) edit distance
• Can we speed things up?• Motivation:
– pages 260-263

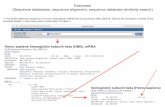
Problem Definition 1
• k-difference global alignment– Input
• Strings S and T– Task
• Find best global alignment of S and T containing at most k mismatches and spaces or say that no such alignment exists

Problem Definition 2
• k-difference inexact matching– Input
• Strings P and T– Task
• Find all ways, if any, to match P in T using at most k character substitutions, insertions, and deletions, or report that no such matches exist.
– End spaces in T but not P are free

• k-mismatch problem– Input
• Strings P and T– Task
• Find all ways, if any, to match P in T using at most k character substitutions, insertions, and deletions, or report that no such matches exist.
– No internal spaces
Earlier Problem Definition

Example
• Difference between k-mismatch problem and the k-difference problem
• Inputs– P = abcdefghi– T = abcdeefghi
• Minimum # of mismatches is 4• Minimum # of differences is 1 with 1 space
in P after the e

Solution for k-difference global alignment
• Compute edit distance of S and T but only fill in an O(km)-size portion of the table
• Work only with diagonals that are within k of the main diagonal
• If result in D(n,m) is <= k, then there is an optimal alignment
• If result in D(n,m) is >k, then the optimal alignment has value > k (though possibly less than D(n,m)

Illustration
01234567890123
0 1 2 3 4 5 6 7 8 9 0 1 2 3
k=4

Unknown k*
• Suppose we don’t know the optimal k* a priori• Use doubling trick to guess k*
– Start with k=1– Then k=2– Then k=4– Then k=8– …– Final work will be O(k*m)

k-difference inexact matching
• Solution method– O(km) time and space solution for the problem
• Can be reduced to O(m+n) space if we only want the end position in T of the match
– Hybrid dynamic programming– Use suffix trees with longest common extension
together with dynamic programming to solve this problem
• Note, the first row of table will be 0 to reflect end spaces in T are free

Definitions
• Diagonals are numbered – 1 to m above main diagonal– -1 to -n below the main diagonal
• A d-path in the dynamic programming table is a path that starts in row 0 and specifies a total of d mismatches and spaces
• A d-path is farthest reaching in diagonal i if – it is a d-path that ends in diagonal i and – its ending column c is >= the ending column of any other
d-path that ends in diagonal i

Illustration 0-1-2-3-4-5-6-7-8-9
1 2 3 4 5 6 7 8 9 0 1 2 3

Approach
• To compute farthest-reaching d-path on diagonal i– Take farthest-reaching (d-1)-path on diagonal i+1
• Move down one square, and then do a longest common extension from that point
– Take farthest-reaching (d-1)-path on diagonal i-1• Move right one square, and then do a longest common
extension from that point
– Take farthest-reaching (d-1)-path on diagonal i• Move diagonally one square, and then do a longest common
extension from that point

Diagonal i+1 (d-1)-path 0-1-2-3-4-5-6-7-8-9
1 2 3 4 5 6 7 8 9 0 1 2 3
Finding longest 1-path on diagonal 3 using longest 0-pathson diagonals 2, 3, and 4 as a starting point.

Diagonal i-1 (d-1)-path 0-1-2-3-4-5-6-7-8-9
1 2 3 4 5 6 7 8 9 0 1 2 3
Finding longest 1-path on diagonal 3 using longest 0-pathson diagonals 2, 3, and 4 as a starting point.

Diagonal i (d-1)-path 0-1-2-3-4-5-6-7-8-9
1 2 3 4 5 6 7 8 9 0 1 2 3
Finding longest 1-path on diagonal 3 using longest 0-pathson diagonals 2, 3, and 4 as a starting point.

High level outline
• d = 0;– For i = 0 to m do
• find longest common extension of P(1) and T(i)• This is the 0-path on diagonal i
• For d = 1 to k do– For i = -n to m do
• using farthest reaching (d-1) paths on diagonals i-1, i, and i+1, find farthest reaching d-path on diagonal i
• Any path that reaches row n defines an inexact match of P in T that contains at most k differences

3. Exclusion Methods
• Previous methods still have running time (km)
• Can we get to expected times of O(m) or even smaller? – Note, we are not asking for worst-case times
this small.• For example, Boyer-Moore has sublinear
time for the exact matching problem

Partition Idea
• Partition T or P into consecutive regions of a given length r
• Search/Filter Phase– Using various exact matching methods, search using
these partition values to filter out possible locations of P in T
• Check phase– For each surviving location, use an approximate
matching technique to verify an approximate occurrence of P

BYP Choices
• Baeza-Yates and Perleberg• O(m) expected running time for modest error rates• Let r = floor(n/(k+1))• Partition P into consecutive length-r intervals
– last interval may have length less than r• Key property
– There are at least k+1 intervals of P that have full length r– If P matches a substring T’ of T with at most k differences,
then T’ must contain one interval of length r that matches one of the k+1 intervals of P exactly.

BYP Algorithm
• Let P’ be the set of k+1 substrings of P taken from the first k+1 regions of P’s partition
• Build a keyword tree for P’• Using Aho-Corasick, find I, the set of all starting
locations in T where any pattern in P occurs exactly• For each i in I, use an approximate matching
algorithm (probably based on dynamic programming) to locate end points of all approximate occurrences of P in substring T[i-n-k..i+n+k]

Running Time Analysis
• Search phase: O(n+m) time and O(n) space– We could use suffix trees or suffix trees and matching
statistics as well for similar performance– We could use Boyer-Moore set matching techniques
described in Section 7.16 to speed this up even more• Check Phase
– Dynamic programming takes O(n2) time per location checked
– Previous results can be used for O(kn) time per location checked

Expected running time
• Need to get expected size of number of locations to be checked
• Probability model– Each character of T is drawn uniformly at random from the
alphabet of size q• An upper bound on the expected number of
occurrences of a region p from P’ in T is m(k+1)/qr
– T has roughly m substrings of length r– Each substring matches an individual p with probability 1/qr

Expected running time
• Expected time of checking: [m(k+1)/qr] n2
– (number of occurrences) x (time per occurrence)• Need to determine what values of k make this
cost <= a constant times m• Some mathematical manipulation leads to BYP
is O(m) as long as k = O(n/log n)– That is, error rate is less than 1 every log n
characters

Extensions
• See the book, pages 273-279, for some extensions to these ideas
• The expected work can be made sublinear