Supplementary Figure and Table - media.nature.com · Supplementary Figure 2. In situ detection of...
Transcript of Supplementary Figure and Table - media.nature.com · Supplementary Figure 2. In situ detection of...

Supplementary Figure 1. The tumors formed in NOD/SCID mice by α2δ1+ and α2δ1– subpopulations isolated from HCC cell lines and primary HCC samples. 1°, primary transplantation; 2°, second transplantation.

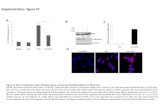
Supplementary Figure 2. In situ detection of let-7c, miR-200b, miR-222 and miR-424 (green), together with DAPI staining (blue) for nuclear in both Hep-11 and Hep-12 cell lines (A), as well as in FACS purified α2δ1- and α2δ1+ Huh7 cells (B). Parallel α2δ1 staining is shown to demonstrate the α2δ1 staining pattern of respective cells. Bar=50 μm.
Supplementary Figure 3. qRT-PCR analysis of indicated miRNA levels in Hep-12 cells (A-D) overexpressing indicated miRNAs, and in Huh7 cells (E-H) with indicated miRNAs knocked-down by Tud RNAs against indicated miRNAs .

Supplementary Figure 4. Uncropped western blots. a, For Fig. 3i; b, For Fig. 3j; c, For Fig. 4c; d, For Fig. 4e.

Supplementary Figure 5. Uncropped western blots. a, For Fig. 5b; b, For Fig. 5d; c, For Fig. 5f; d, For Fig. 5h.

Supplementary Figure 6. Uncropped western blots. a, For Fig. 6f; b, For Fig. 6g; c, For Fig. 6n

Supplementary Figure 7. Uncropped western blots. a, For Fig.7a; b, For Fig. 7c; c, For Fig. 7e; d, For Fig. 7g; e, For Fig. 7h; f, For Fig. 7i.

Supplementary Table 1. Relationships between the expression of α2δ1 mRNA
and the clinicopathologic features in 85 HCC patients
Variable Cases
(n)
α2δ1 expression
(RQ: 2- Ct) P-value a
Median Range
Gender Male 74 1.26 0.003-46.45- 0.465
Female 11 1.39 0.29-161.99
Age (year) ≤60 63 1.30 0.003-46.45 0.256
>60 22 1.12 0.21-161.99
Cirrhosis Absent 27 0.84 0.003-15.69 0.728
Present 58 1.53 0.21-161.99
Size (cm) ≤5 45 0.82 0.003-39.31 0.030
>5 40 1.94 0.25-161.99
Venous invasion Absent 63 1.27 0.003-161.99 0.173
Present 22 1.56 0.26-46.45
Survival (year) <4 40 1.777 0.003-161.99 0.046
≥4 45 1.187 0.21-8.693
Recurrence Yes b 41 1.87 0.003-161.99 0.0096
No c 44 0.99 0.09-15.69 aMann–Whitney test. bRecurrences occur within 2 years of diagnosis. cNo recurrences found within 4 years of diagnosis.

Supplementary Table 2. Cox regression analysis of clinicopathological features and α2δ1 expression in 85 HCC patients
Variable Cases
(n)
Multivariate analysisa
RR(95%CI) P-value
Sex Male 74 1.379 0.653
Female 11 0.565-3.363
Age (year) ≤60 63 1.380 0.281
>60 22 0.641–2.972
Cirrhosis Absent 27 0.502 0.023
Present 58 0.231–1.091
Size (cm) ≤5 45 7.044 0.009
>5 40 2.660-18.665
Venous invasion Present 22 1.562 0.024
Absent 63 1.155-2.113
α2δ1 expressionb High 43 2.662 0.005
Low 42 1.355-5.232
a COX regression analysis.
bThe patients are classified according to the median level of α2δ1 expression.
Abbreviations: RR, Relative risk; CI, Confidence interval.

Supplementary Table 3. The relationships between the expression of PBX3 mRNA
and the clinicopathologic features in 85 HCC patients
Variable Cases
(n)
PBX3 expression (RQ: 2- Ct) P-value a
Median Range
Gender Male 74 2.51 0.01-62850 0.260
Female 11 3.72 0.10-46.51
Age (year) ≤60 63 3.16 0.01-371.6 0.474
>60 22 3.01 0.06-62850
Cirrhosis Absent 27 1.51 0.01-56.43 0.028
Present 58 3.83 0.06-62850
Size (cm) ≤5 45 1.51 0.01-5535 0.038
>5 40 4.48 0.06-62850
Venous invasion Absent 63 2.67 0.01-62850 0.313
Present 22 2.30 0.06-371.6
Survival (year) <4 40 3.94 0.01-62850 0.024
≥4 45 2.16 0.06-138.3
Recurrence Yes b 41 3.38 0.09-62850 0.0074
No c 44 1.56 0.01-53.26 aMann–Whitney test. bRecurrences occur within 2 years of diagnosis. cNo recurrences detected within 4 years of diagnosis.

Supplementary Table 4. Cox regression analysis of clinicopathological features and
PBX3 expression in 85 HCC patients
Variable Cases(n)
Multivariate analysisa
RR(95%CI) P-value
Sex Male 74 0.824 0.668
Female 11 0.339 - 2.000
Age (year) ≤60 63 1.493 0.368
>60 22 0.633 - 3.519
Cirrhosis Absent 27 0.463 0.102
Present 58 0.184 - 1.116
Size (cm) ≤5 45 2.406 0.035
>5 40 1.065 - 5.434
Venous invasion Absent 63 0.318 0.005
Present 22 0.143 - 0.707
PBX3b High 44 2.935 0.008
Low 41 1.329 - 6.483
a COX regression analysis.
bThe patients are classified according to the median level of PBX3 mRNA.
Abbreviation: RR: Relative risk, CI: Confidence interval

Supplementary Table 5. PBX3 responsive genes as revealed by RNA‐seq analysis of SMMC‐7721 cells
upon forced expression of PBX3 compared with vector control
Up‐regulated genes after PBX3 over‐expression
Gene Symbol Gene name Ratio
BG alpha‐1‐B glycoprotein 41.51
ABCB1 ATP‐binding cassette, sub‐family B (MDR/TAP), member 1 3.89
ABHD1 abhydrolase domain containing 1 2.83
ABI1 abl‐interactor 1 2.64
ABI3 ABI family, member 3 17.82
ACOT6 acyl‐CoA thioesterase 6 2.64
ACSL6 acyl‐CoA synthetase long‐chain family member 6 8.24
ACSM2A acyl‐CoA synthetase medium‐chain family member 2A 67.83
ADAM28 ADAM metallopeptidase domain 28 95.57
ADCY2 adenylate cyclase 2 (brain) 30.73
ADPGK ADP‐dependent glucokinase 31.65
AHSP erythroid associated factor 476.19
AKR1B15 aldo‐keto reductase family 1, member B10 (aldose reductase) 32.25
ALDOC aldolase C, fructose‐bisphosphate 3.11
ALOX12B arachidonate 12‐lipoxygenase, 12R type 2.59
ALOX5 arachidonate 5‐lipoxygenase 16.66
ALOXE3 arachidonate lipoxygenase 3 28.09
AMBP alpha‐1‐microglobulin/bikunin precursor 2.19
AMN amnionless homolog (mouse) 32.11
ANGPTL3 angiopoietin‐like 3 254.27
ANK1 ankyrin 1, erythrocytic 10.05
ANK2 ankyrin 2, neuronal 16.88
ANKRD12 ankyrin repeat domain 12 2.45
ANKRD22 ankyrin repeat domain 22 2.23
ANKRD45 ankyrin repeat domain 45 19.49
ANO2 anoctamin 2 10.38
APCDD1L adenomatosis polyposis coli down‐regulated 1‐like 29.48
APLN apelin 2.47
APOA1 apolipoprotein A‐I 41.18
APOBEC2 apolipoprotein B mRNA editing enzyme, catalytic polypeptide‐like 2 40.57
ARHGAP15 Rho GTPase activating protein 15 19.52
ARHGAP19 Rho GTPase activating protein 19 11.40
ARHGAP4 Rho GTPase activating protein 4 3.36
ARL17B ADP‐ribosylation factor‐like 17 pseudogene 1 2.09
ARR3 arrestin 3, retinal (X‐arrestin) 107.00
ARSD arylsulfatase D 27.50
ART1 ADP‐ribosyltransferase 1 150.86
ASGR1 asialoglycoprotein receptor 1 37.87
ASPN asporin 79.26
ATF3 activating transcription factor 3 3.05

ATG9B ATG9 autophagy related 9 homolog B (S. cerevisiae) 2.67
ATP6V0A4 ATPase, H+ transporting, lysosomal V0 subunit a4 29.29
ATXN3 ataxin 3 2.16
B3GNT8 UDP‐GlcNAc:betaGal beta‐1,3‐N‐acetylglucosaminyltransferase 8 4.04
BCL2L14 BCL2‐like 14 (apoptosis facilitator) 98.77
BCL2L2 BCL2‐like 2 2.14
BEST2 bestrophin 2 148.04
BEST4 bestrophin 4 2.90
BMP5 bone morphogenetic protein 5 10.17
BNIP3 BCL2/adenovirus E1B 19kDa interacting protein 3 3.65
BOC Boc homolog (mouse) 17.75
C10orf10 chromosome 10 open reading frame 10 2.41
C11orf16 chromosome 11 open reading frame 16 18.00
C15orf26 chromosome 15 open reading frame 26 126.19
C16orf11 hypothetical protein FLJ36208; chromosome 16 open reading frame
11
89.71
C1orf116 chromosome 1 open reading frame 116 10.60
C1orf21 chromosome 1 open reading frame 21 2.61
C1orf228 chromosome 1 open reading frame 228 2.04
C2 complement component 2 3.21
C2orf72 chromosome 2 open reading frame 72 34.96
C2orf74 chromosome 2 open reading frame 74 4.56
C2orf78 chromosome 2 open reading frame 78 31.61
C6orf163 chromosome 6 open reading frame 163 2.59
C6orf201 chromosome 6 open reading frame 201 3.74
C7 complement component 7 12.95
C7orf65 chromosome 7 open reading frame 65 27.82
C8orf74 chromosome 8 open reading frame 74 294.85
C8orf86 chromosome 8 open reading frame 86 167.46
C9orf50 chromosome 9 open reading frame 50 151.89
CA9 carbonic anhydrase IX 5.56
CABP7 calcium binding protein 7 2.36
CACNA2D1 calcium channel, voltage‐dependent, alpha 2/delta subunit 1 2884.80
CAMK4 calcium/calmodulin‐dependent protein kinase IV 11.77
CAPN5 calpain 5 4.75
CARD18 caspase recruitment domain family, member 18 14.45
CASC1 cancer susceptibility candidate 1 26.48
CASQ1 calsequestrin 1 (fast‐twitch, skeletal muscle) 2.16
CCDC105 coiled‐coil domain containing 105 115.02
CCDC106 coiled‐coil domain containing 106 2.46
CCDC114 coiled‐coil domain containing 114 3.61
CCDC122 coiled‐coil domain containing 122 2.66
CCDC149 coiled‐coil domain containing 149 3.37
CCDC17 coiled‐coil domain containing 17 15.76

CCDC33 coiled‐coil domain containing 33 37.22
CCDC62 coiled‐coil domain containing 62 2.58
CD70 CD70 molecule 2.12
CD79A CD79a molecule, immunoglobulin‐associated alpha 16.39
CDK18 PCTAIRE protein kinase 3 2.52
CEACAM16 carcinoembryonic antigen‐related cell adhesion molecule 16 188.20
CEACAM19 carcinoembryonic antigen‐related cell adhesion molecule 19 15.11
CES3 carboxylesterase 3 9.76
CHD5 chromodomain helicase DNA binding protein 5 2.26
CHRNB4 cholinergic receptor, nicotinic, beta 4 24.77
CHRNG cholinergic receptor, nicotinic, gamma 32.30
CIB2 calcium and integrin binding family member 2 27.21
CISH cytokine inducible SH2‐containing protein 19.48
CLCA2 chloride channel accessory 2 7.00
CLDN7 claudin 7 3.75
CLEC14A C‐type lectin domain family 14, member A 16.89
CLGN calmegin 2.11
CLIC3 chloride intracellular channel 3 2.52
CLIP3 CAP‐GLY domain containing linker protein 3 2.81
CLUL1 clusterin‐like 1 (retinal) 13.16
CNIH3 cornichon homolog 3 (Drosophila) 19.49
CNTNAP2 contactin associated protein‐like 2 3.05
COL13A1 collagen, type XIII, alpha 1 35.02
COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) 4.81
CPLX3 complexin 3 71.78
CPNE5 copine V 73.87
CPXM2 carboxypeptidase X (M14 family), member 2 3.41
CRYGS crystallin, gamma S 47.87
CSTA cystatin A (stefin A) 5.54
CTSW cathepsin W 26.06
CXCR5 chemokine (C‐X‐C motif) receptor 5 15.22
CYP39A1 cytochrome P450, family 39, subfamily A, polypeptide 1 2.86
CYP4X1 cytochrome P450, family 4, subfamily X, polypeptide 1 59.79
CYP8B1 cytochrome P450, family 8, subfamily B, polypeptide 1 50.99
DBNDD1 dysbindin (dystrobrevin binding protein 1) domain containing 1 17.44
DGKI diacylglycerol kinase, iota 9.37
DHDH dihydrodiol dehydrogenase (dimeric) 22.65
DHH desert hedgehog homolog (Drosophila) 27.78
DHRS9 dehydrogenase/reductase (SDR family) member 9 7.25
DIRAS1 DIRAS family, GTP‐binding RAS‐like 1 7.16
DNAJB5 DnaJ (Hsp40) homolog, subfamily B, member 5 2.25
DNAJC28 DnaJ (Hsp40) homolog, subfamily C, member 28 2.23
DNALI1 dynein, axonemal, light intermediate chain 1 22.53
DPCR1 diffuse panbronchiolitis critical region 1 15.96

DPPA4 developmental pluripotency associated 4 11.40
DSE dermatan sulfate epimerase 2.70
DUOX2 dual oxidase 2 2.16
EFEMP2 EGF‐containing fibulin‐like extracellular matrix protein 2 5.30
EFHD1 EF‐hand domain family, member D1 18.39
EGFL8 EGF‐like‐domain, multiple 8 2.44
EIF4ENIF1 eukaryotic translation initiation factor 4E nuclear import factor 1 2.10
ELAVL3 ELAV (embryonic lethal, abnormal vision, Drosophila)‐like 3 (Hu
antigen C)
30.49
ELOVL7 ELOVL family member 7, elongation of long chain fatty acids (yeast) 2.24
ELTD1 EGF, latrophilin and seven transmembrane domain containing 1 17.58
EMILIN3 elastin microfibril interfacer 3 2.95
EMR1 egf‐like module containing, mucin‐like, hormone receptor‐like 1 12.87
ENKUR enkurin, TRPC channel interacting protein 34.55
ENPP7 ectonucleotide pyrophosphatase/phosphodiesterase 7 32.87
EPCAM epithelial cell adhesion molecule 24.20
EPN2 epsin 2 2.88
ESF1 similar to ABT1‐associated protein; ESF1, nucleolar pre‐rRNA
processing protein, homolog (S. cerevisiae)
11.30
EXTL1 exostoses (multiple)‐like 1 15.88
EYA2 eyes absent homolog 2 (Drosophila) 77.76
FAM109B family with sequence similarity 109, member B 13.31
FAM117A family with sequence similarity 117, member A 3.84
FAM180A family with sequence similarity 180, member A 282.44
FAM196A chromosome 10 open reading frame 141 45.70
FAM19A3 family with sequence similarity 19 (chemokine (C‐C motif)‐like),
member A3
24.31
FAM25C family with sequence similarity 25, member C 300.60
FAM92B family with sequence similarity 92, member B 106.17
FBLN2 fibulin 2 19.19
FBXO24 F‐box protein 24 17.78
FCGR1A Fc fragment of IgG, high affinity Ic, receptor (CD64) 13.19
FCRL6 Fc receptor‐like 6 18.65
FCRLB Fc receptor‐like B 5.29
FER1L5 fer‐1‐like 5 (C. elegans) 25.03
FER1L6 fer‐1‐like 6 (C. elegans) 47.07
FFAR1 free fatty acid receptor 1 19.14
FGD2 FYVE, RhoGEF and PH domain containing 2 22.35
FGD5 FYVE, RhoGEF and PH domain containing 5 108.00
FGF22 fibroblast growth factor 22 2.64
FGGY FGGY carbohydrate kinase domain containing 32.46
FKBP1B FK506 binding protein 1B, 12.6 kDa 18.24
FLG2 filaggrin family member 2 10.52
FLJ22184 hypothetical protein FLJ22184 2.25

FLJ44635 TPT1‐like protein 17.26
FMN1 formin 1 15.46
FOXD4L1 forkhead box D4‐like 1 40.44
FOXS1 forkhead box S1 148.86
FRK fyn‐related kinase 2.10
FRMPD4 FERM and PDZ domain containing 4 6.22
FSIP2 fibrous sheath interacting protein 2 13.29
FSTL4 follistatin‐like 4 8.83
FUZ fuzzy homolog (Drosophila) 2.41
GAB2 GRB2‐associated binding protein 2 2.75
GABBR2 gamma‐aminobutyric acid (GABA) B receptor, 2 8.50
GAL3ST1 galactose‐3‐O‐sulfotransferase 1 128.88
GALNT12 UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide
N‐acetylgalactosaminyltransferase 12 (GalNAc‐T12)
4.15
GALNT8 UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide
N‐acetylgalactosaminyltransferase 8 (GalNAc‐T8)
94.33
GEM GTP binding protein overexpressed in skeletal muscle 112.38
GIMAP1 GTPase, IMAP family member 1 11.28
GIMAP8 GTPase, IMAP family member 8 22.65
GLIS1 GLIS family zinc finger 1 21.91
GNGT1 guanine nucleotide binding protein (G protein), gamma transducing
activity polypeptide 1
49.22
GNRHR gonadotropin‐releasing hormone receptor 9.92
GOLGA6D golgi autoantigen, golgin subfamily a, 6D 12.41
GOLGA6L6 Putative golgin subfamily A member 6‐like protein 6 10.76
GPA33 glycoprotein A33 (transmembrane) 22.00
GPIHBP1 glycosylphosphatidylinositol anchored high density lipoprotein
binding protein 1
19.81
GPNMB glycoprotein (transmembrane) nmb 2.25
GPR155 G protein‐coupled receptor 155 33.09
GPR158 G protein‐coupled receptor 158 11.81
GPR176 G protein‐coupled receptor 176 12.62
GPR85 G protein‐coupled receptor 85 29.09
GRAMD3 GRAM domain containing 3 2.13
GRIK2 glutamate receptor, ionotropic, kainate 2 4.20
GRIP2 glutamate receptor interacting protein 2 2.75
GSTA2 glutathione S‐transferase alpha 2 152.58
GULP1 GULP, engulfment adaptor PTB domain containing 1 2.90
HAAO 3‐hydroxyanthranilate 3,4‐dioxygenase 154.80
HAPLN2 hyaluronan and proteoglycan link protein 2 2.64
HIC1 hypermethylated in cancer 1 24.58
HIF1A hypoxia inducible factor 1, alpha subunit (basic helix‐loop‐helix
transcription factor)
16.95
HIST1H3J histone cluster 1, H3j 2.00

HIST1H4A histone cluster 1, H4a 19.39
HLA‐DRB4 major histocompatibility complex, class II, DR beta 4 16.83
HOXC12 homeobox C12 290.54
HOXD3 homeobox D3 2.29
HS3ST3B1 heparan sulfate (glucosamine) 3‐O‐sulfotransferase 3B1 2.05
HSPB2 heat shock 27kDa protein 2 28.07
HSPB7 heat shock 27kDa protein family, member 7 (cardiovascular) 64.69
IFNA8 interferon, alpha 8 18.04
IGFBP2 insulin‐like growth factor binding protein 2, 36kDa 197.93
IGFBP5 insulin‐like growth factor binding protein 5 14.42
IL17D interleukin 17D 3.58
IL1RAP interleukin 1 receptor accessory protein 33.89
INHBC inhibin, beta C 2.02
INMT indolethylamine N‐methyltransferase 13.90
INSM2 insulinoma‐associated 2 3.05
IQUB IQ motif and ubiquitin domain containing 46.59
ISPD notch1‐induced protein 3.24
ITGAL integrin, alpha L (antigen CD11A (p180), lymphocyte
function‐associated antigen 1; alpha polypeptide)
13.93
ITGBL1 integrin, beta‐like 1 (with EGF‐like repeat domains) 21.97
JAZF1 JAZF zinc finger 1 32.89
JSRP1 junctional sarcoplasmic reticulum protein 1 6.48
KAT6B K(lysine) acetyltransferase 6B 2.23
KATNAL1 katanin p60 subunit A‐like 1 2.63
KCNB1 potassium voltage‐gated channel, Shab‐related subfamily, member 1 131.34
KCNC1 potassium voltage‐gated channel, Shaw‐related subfamily, member 1 43.60
KCND2 potassium voltage‐gated channel, Shal‐related subfamily, member 2 15.92
KCNE3 potassium voltage‐gated channel, Isk‐related family, member 3 14.84
KCNJ14 potassium inwardly‐rectifying channel, subfamily J, member 14 2.12
KCNK2 potassium channel, subfamily K, member 2 19.87
KCNQ5 potassium voltage‐gated channel, KQT‐like subfamily, member 5 11.18
KCNS1 potassium voltage‐gated channel, delayed‐rectifier, subfamily S,
member 1
32.23
KCTD12 potassium channel tetramerisation domain containing 12 2.29
KIAA1045 KIAA1045 15.08
KIAA1324 KIAA1324 7.80
KIAA1462 KIAA1462 9.85
KIAA1683 KIAA1683 2.34
KIF26B kinesin family member 26B 19.26
KIRREL3 kin of IRRE like 3 (Drosophila) 3.24
KLHL32 kelch‐like 32 (Drosophila) 6.06
KLHL7 kelch‐like 7 (Drosophila) 2.14
KLRF1 killer cell lectin‐like receptor subfamily F, member 1 40.41
KRT20 keratin 20 2.41

KRT32 keratin 32 426.17
KRT78 keratin 78 19.46
LAIR1 leukocyte‐associated immunoglobulin‐like receptor 1 11.71
LEPR leptin receptor 15.92
LGALS4 lectin, galactoside‐binding, soluble, 4 540.41
LHB luteinizing hormone beta polypeptide 25.42
LHCGR luteinizing hormone/choriogonadotropin receptor 14.78
LIMS2 LIM and senescent cell antigen‐like domains 2 47.15
LIN28A lin‐28 homolog (C. elegans) 10.45
LIPC lipase, hepatic 28.94
LIPN lipase, family member N 2.86
LOC100129924 similar to hCG2036949 2.44
LOC100287036 hypothetical protein LOC100287036 6.17
LOC388813 uncharacterized protein ENSP00000383407‐like 226.55
LOC554223 hypothetical LOC554223 86.05
LOC728392 hypothetical protein LOC728392 2.75
LOXL4 lysyl oxidase‐like 4 7.86
LRRC17 leucine rich repeat containing 17 14.84
LRRC25 leucine rich repeat containing 25 4.83
LRRC39 leucine rich repeat containing 39 20.81
LRRC4 leucine rich repeat containing 4 26.95
LY6D lymphocyte antigen 6 complex, locus D 28.96
MAF v‐maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) 47.09
MAGI2 membrane associated guanylate kinase, WW and PDZ domain
containing 2
4.51
MANSC1 MANSC domain containing 1 2.83
MAP7D2 MAP7 domain containing 2 10.41
MARCO macrophage receptor with collagenous structure 99.25
MAT1A methionine adenosyltransferase I, alpha 28.12
MB myoglobin 3.06
MED12L mediator complex subunit 12‐like 4.51
MFSD2B hypothetical protein LOC388931 163.57
MLIP muscular LMNA‐interacting protein 30.43
MMP23B matrix metallopeptidase 23A (pseudogene); matrix metallopeptidase
23B
2.31
MRAP2 melanocortin 2 receptor accessory protein 2 7.37
MSI1 musashi homolog 1 (Drosophila) 67.82
MTL5 metallothionein‐like 5, testis‐specific (tesmin) 3.79
MXD1 MAX dimerization protein 1 11.17
MYBPC2 myosin binding protein C, fast type 39.58
MYBPC3 myosin binding protein C, cardiac 3.24
MYLK3 myosin light chain kinase 3 2.67
MYO1A myosin IA 13.60
MYOZ1 myozenin 1 53.69

NEGR1 neuronal growth regulator 1 3.43
NEU2 sialidase 2 (cytosolic sialidase) 768.05
NGFR nerve growth factor receptor (TNFR superfamily, member 16) 29.83
NHLRC4 hypothetical protein FLJ36208; chromosome 16 open reading frame
11
3.05
NKD2 naked cuticle homolog 2 (Drosophila) 14.87
NOL3 nucleolar protein 3 (apoptosis repressor with CARD domain) 2.21
NOS3 nitric oxide synthase 3 (endothelial cell) 4.36
NOSTRIN nitric oxide synthase trafficker 25.09
NOTCH3 Notch homolog 3 (Drosophila) 17.18
NOVA2 neuro‐oncological ventral antigen 2 185.06
NPL N‐acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) 11.92
NPW neuropeptide W 2.16
NPY5R neuropeptide Y receptor Y5 73.56
NR5A1 nuclear receptor subfamily 5, group A, member 1 14.78
NR5A2 nuclear receptor subfamily 5, group A, member 2 30.12
NTF4 neurotrophin 4 450.80
NTNG2 netrin G2 20.84
NXNL2 nucleoredoxin‐like 2 3.83
NYNRIN KIAA1305 38.45
ODAM odontogenic, ameloblast asssociated 55.84
OLFM2 olfactomedin 2 29.83
OMD osteomodulin 112.05
OMG oligodendrocyte myelin glycoprotein 2.07
OR13A1 olfactory receptor, family 13, subfamily A, member 1 12.75
OR4F21 olfactory receptor, family 4, subfamily F, member 21 26.83
OR6K3 olfactory receptor, family 6, subfamily K, member 3 26.71
ORAI3 ORAI calcium release‐activated calcium modulator 3 2.29
OTOGL otogelin‐like 24.92
OVOL1 ovo‐like 1(Drosophila) 2.16
P2RX3 purinergic receptor P2X, ligand‐gated ion channel, 3 60.93
P4HA1 prolyl 4‐hydroxylase, alpha polypeptide I 8.17
PAK4 p21 protein (Cdc42/Rac)‐activated kinase 4 2.31
PALM3 Paralemmin‐3 50.43
PAPPA2 pappalysin 2 46.30
PARK2 Parkinson disease (autosomal recessive, juvenile) 2, parkin 9.27
PBOV1 prostate and breast cancer overexpressed 1 12.94
PBX3 pre‐B‐cell leukemia homeobox 3 88.94
PCDHA9 protocadherin alpha 9 2.16
PCOLCE procollagen C‐endopeptidase enhancer 211.29
PDE6G phosphodiesterase 6G, cGMP‐specific, rod, gamma 21.42
PDGFC platelet derived growth factor C 5.64
PDIA2 protein disulfide isomerase family A, member 2 233.37
PDK3 pyruvate dehydrogenase kinase, isozyme 3 2.65

PDLIM4 PDZ and LIM domain 4 19.77
PIWIL4 piwi‐like 4 (Drosophila) 20.51
PKD1L1 polycystic kidney disease 1 like 1 14.95
PLA2G4D phospholipase A2, group IVD (cytosolic) 130.30
PLAG1 pleiomorphic adenoma gene 1 18.21
PLAT plasminogen activator, tissue 5.04
PLEKHA4 pleckstrin homology domain containing, family A (phosphoinositide
binding specific) member 4
5.49
PLXDC1 plexin domain containing 1 2.58
PLXNA4 plexin A4 39.31
POPDC2 popeye domain containing 2 2.75
PPFIA1 protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF),
interacting protein (liprin), alpha 1
2.03
PPFIA4 protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF),
interacting protein (liprin), alpha 4
4.41
PPP1R1C protein phosphatase 1, regulatory (inhibitor) subunit 1C 223.65
PPP1R3C protein phosphatase 1, regulatory (inhibitor) subunit 3C 2.77
PRELID2 PRELI domain containing 2 3.10
PRKAG2 protein kinase, AMP‐activated, gamma 2 non‐catalytic subunit 2.52
PRKAR2B protein kinase, cAMP‐dependent, regulatory, type II, beta 35.87
PRND prion protein 2 (dublet) 2.52
PRPH peripherin 55.44
PRR23C FLJ46210 protein 23.35
PRSS16 protease, serine, 16 (thymus) 2.11
PTAFR platelet‐activating factor receptor 11.49
PTPRC protein tyrosine phosphatase, receptor type, C 12.89
PTPRE protein tyrosine phosphatase, receptor type, E 2.37
PTPRH protein tyrosine phosphatase, receptor type, H 3.09
PTPRR protein tyrosine phosphatase, receptor type, R 54.15
RAB43 RAB43, member RAS oncogene family; hypothetical LOC100131426 18.55
RAG1 recombination activating gene 1 3.74
RAPGEF5 Rap guanine nucleotide exchange factor (GEF) 5 30.38
RARRES1 retinoic acid receptor responder (tazarotene induced) 1 33.08
RASSF4 Ras association (RalGDS/AF‐6) domain family member 4 4.32
RBPJL recombination signal binding protein for immunoglobulin kappa J
region‐like
27.77
RCN3 reticulocalbin 3, EF‐hand calcium binding domain 3.87
RDX radixin 2.01
RFESD Rieske (Fe‐S) domain containing 2.48
RFX3 regulatory factor X, 3 (influences HLA class II expression) 23.46
RGAG1 retrotransposon gag domain containing 1 3.24
RGMA RGM domain family, member A 18.12
RIN2 Ras and Rab interactor 2 11.91
RNASEK ribonuclease, RNase K 21.06

RNF182 ring finger protein 182 2.13
RNF222 ring finger protein 222 2.07
ROS1 c‐ros oncogene 1 , receptor tyrosine kinase 2.11
RPS6KB1 ribosomal protein S6 kinase, 70kDa, polypeptide 1 2.21
RTDR1 rhabdoid tumor deletion region gene 1 39.71
RTN4RL2 reticulon 4 receptor‐like 2 3.52
SALL2 sal‐like 2 (Drosophila) 6.24
SCARF2 scavenger receptor class F, member 2 12.00
SCG5 secretogranin V (7B2 protein) 36.45
SCGB1D2 secretoglobin, family 1D, member 2 627.37
SCN2B sodium channel, voltage‐gated, type II, beta 8.43
SCT secretin 479.89
SDC2 syndecan 2 6.96
SEMA5A sema domain, seven thrombospondin repeats (type 1 and type
1‐like), transmembrane domain (TM) and short cytoplasmic domain,
(semaphorin) 5A
2.44
SERPINC1 serpin peptidase inhibitor, clade C (antithrombin), member 1 31.09
SERPINE3 serpin peptidase inhibitor, clade E (nexin, plasminogen activator
inhibitor type 1), member 3
21.66
SERPINF2 serpin peptidase inhibitor, clade F (alpha‐2 antiplasmin, pigment
epithelium derived factor), member 2
17.57
SFMBT2 Scm‐like with four mbt domains 2 9.62
SGK2 serum/glucocorticoid regulated kinase 2 20.06
SGOL1 shugoshin‐like 1 (S. pombe) 9.40
SGOL2 shugoshin‐like 2 (S. pombe) 12.89
SH3YL1 SH3 domain containing, Ysc84‐like 1 (S. cerevisiae) 3.05
SHISA7 UPF0626 protein A 10.58
SIAH3 seven in absentia homolog 3 (Drosophila) 35.56
SIGIRR single immunoglobulin and toll‐interleukin 1 receptor (TIR) domain 14.52
SLC14A2 solute carrier family 14 (urea transporter), member 2 9.59
SLC22A11 solute carrier family 22 (organic anion/urate transporter), member
11
111.92
SLC22A14 solute carrier family 22, member 14 36.99
SLC2A14 solute carrier family 2 (facilitated glucose transporter), member 14 35.10
SLC2A5 solute carrier family 2 (facilitated glucose/fructose transporter),
member 5
2.06
SLC37A2 solute carrier family 37 (glycerol‐3‐phosphate transporter), member
2
13.93
SLC43A1 solute carrier family 43, member 1 44.14
SLC5A2 solute carrier family 5 (sodium/glucose cotransporter), member 2 24.36
SLC6A8 solute carrier family 6 (neurotransmitter transporter, creatine),
member 8
63.59
SLC8A2 solute carrier family 8 (sodium/calcium exchanger), member 2 32.53
SLCO1A2 solute carrier organic anion transporter family, member 1A2 7.14

SLCO1B1 solute carrier organic anion transporter family, member 1B1 101.72
SLCO1B7 solute carrier organic anion transporter family, member 1B7 4.04
SMARCD3 SWI/SNF related, matrix associated, actin dependent regulator of
chromatin, subfamily d, member 3
2.28
SMTN smoothelin 3.53
SNX20 sorting nexin 20 43.10
SOX2 SRY (sex determining region Y)‐box 2 3.74
SPACA4 sperm acrosome associated 4 26.29
SPTY2D1 SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) 24.41
SRCRB4D scavenger receptor cysteine rich domain containing, group B (4
domains)
2.16
SRMS src‐related kinase lacking C‐terminal regulatory tyrosine and
N‐terminal myristylation sites
4.95
STX1A syntaxin 1A (brain) 19.62
SULT1A2 sulfotransferase family, cytosolic, 1A, phenol‐preferring, member 2 2.22
SWT1 Transcriptional protein SWT1 11.15
SYCP2 synaptonemal complex protein 2 2.00
SYCP2L synaptonemal complex protein 2‐like 6.67
SYN2 synapsin II 2.64
TACR1 tachykinin receptor 1 51.44
TAF7L TAF7‐like RNA polymerase II, TATA box binding protein
(TBP)‐associated factor, 50kDa
16.89
TAS2R5 taste receptor, type 2, member 5 2.25
TBC1D3C TBC1 domain family, member 3C 22.31
TBX19 T‐box 19 2.14
TCEAL3 transcription elongation factor A (SII)‐like 3 3.26
TCTE3 t‐complex‐associated‐testis‐expressed 3 54.93
TEK TEK tyrosine kinase, endothelial 25.59
TFEC transcription factor EC 8.70
THY1 Thy‐1 cell surface antigen 16.15
TICAM2 transmembrane emp24 protein transport domain containing 7;
toll‐like receptor adaptor molecule 2
30.55
TLR9 toll‐like receptor 9 32.16
TM4SF19 transmembrane 4 L six family member 19 2.80
TMEM119 transmembrane protein 119 52.73
TMEM191C transmembrane protein 191B; transmembrane protein 191C 34.32
TMEM63C transmembrane protein 63C 2.34
TMEM88 transmembrane protein 88 34.92
TNFRSF13B tumor necrosis factor receptor superfamily, member 13B 547.26
TOX thymocyte selection‐associated high mobility group box 168.89
TP53INP2 tumor protein p53 inducible nuclear protein 2 2.13
TP53TG5 TP53 target 5 18.43
TRAF5 TNF receptor‐associated factor 5 3.54
TREM1 triggering receptor expressed on myeloid cells 1 466.94

TRIM39 tripartite motif‐containing 39 16.63
TRIM63 tripartite motif‐containing 63 197.36
TRIM71 tripartite motif‐containing 71 78.61
TSHZ2 teashirt zinc finger homeobox 2 7.55
TSPAN5 tetraspanin 5 3.51
TSPAN8 tetraspanin 8 3.29
UBE2W ubiquitin‐conjugating enzyme E2W (putative) 7.36
UBQLNL ubiquilin‐like 16.96
UCHL1 ubiquitin carboxyl‐terminal esterase L1 (ubiquitin thiolesterase) 44.44
UGT2B15 UDP glucuronosyltransferase 2 family, polypeptide B15 27.32
UGT2B17 UDP glucuronosyltransferase 2 family, polypeptide B17 19.99
USHBP1 Usher syndrome 1C binding protein 1 16.75
VRTN Vertebrae Development Homolog 2.16
WFIKKN1 WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain
containing 1
12.80
WNT10A wingless‐type MMTV integration site family, member 10A 189.62
WTH3DI RAB6C‐like 3.40
ZAP70 zeta‐chain (TCR) associated protein kinase 70kDa 33.22
ZCCHC12 zinc finger, CCHC domain containing 12 24.72
ZCCHC16 zinc finger, CCHC domain containing 16 14.47
ZDHHC22 zinc finger, DHHC‐type containing 22 41.79
ZFHX4 zinc finger homeobox 4 27.12
ZFP2 zinc finger protein 2 homolog (mouse) 104.71
ZFR2 zinc finger RNA binding protein 2 43.64
ZNF438 zinc finger protein 438 14.83
ZNF474 zinc finger protein 474 3.05
ZNF521 zinc finger protein 521 78.12
ZNF649 zinc finger protein 649 14.46
ZNF674 zinc finger family member 674 2.05
ZP3 zona pellucida glycoprotein 3 (sperm receptor) 2.84
ZSCAN30 zinc finger and SCAN domain containing 30 2.19
Down‐regulated genes following PBX3 overexpression
Gene Symbol Gene name Ratio
ACSBG1 acyl‐CoA synthetase bubblegum family member 1 0.272
ACSM3 acyl‐CoA synthetase medium‐chain family member 3 0.024
ACTL8 actin‐like 8 0.472
ADAM8 ADAM metallopeptidase domain 8 0.106
ADAMTSL2 similar to ADAMTS‐like 2; ADAMTS‐like 2 0.398
ADAP1 ArfGAP with dual PH domains 1 0.452
ADAP2 ArfGAP with dual PH domains 2 0.050
ADC arginine decarboxylase 0.040
ADD3 adducin 3 (gamma) 0.265
ADRA1D adrenergic, alpha‐1D‐, receptor 0.005
AIM2 absent in melanoma 2 0.052

AIRE autoimmune regulator 0.019
AMACR C1q and tumor necrosis factor related protein 3;
alpha‐methylacyl‐CoA racemase 0.072
AMMECR1 Alport syndrome, mental retardation, midface hypoplasia and
elliptocytosis chromosomal region gene 1 0.110
ANGPTL5 angiopoietin‐like 5 0.341
ANKRD35 ankyrin repeat domain 35 0.025
AP3B2 adaptor‐related protein complex 3, beta 2 subunit 0.257
APOF apolipoprotein F 0.046
APOL3 apolipoprotein L, 3 0.130
APOL6 apolipoprotein L, 6 0.391
APOM apolipoprotein M 0.375
AREG amphiregulin; amphiregulin B 0.483
ARID3C AT rich interactive domain 3C (BRIGHT‐like) 0.036
ARMCX2 armadillo repeat containing, X‐linked 2 0.080
ARRB1 arrestin, beta 1 0.356
ASB14 ankyrin repeat and SOCS box‐containing 14 0.099
ATP2A1 ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 0.092
ATP5J2‐PTCD1 ATP5J2‐PTCD1 Read‐Through Transcript 0.105
ATP5SL ATP5S‐like 0.073
ATP8B4 ATPase, class I, type 8B, member 4 0.090
BCL2L11 BCL2‐like 11 (apoptosis facilitator) 0.307
BCMO1 beta‐carotene 15,15'‐monooxygenase 1 0.008
BDH1 3‐hydroxybutyrate dehydrogenase, type 1 0.324
BST1 bone marrow stromal cell antigen 1 0.048
BST2 NPC‐A‐7; bone marrow stromal cell antigen 2 0.444
BTBD16 BTB (POZ) domain containing 16 0.048
BTBD18 hypothetical protein LOC643376 0.061
BTC betacellulin 0.034
C12orf61 chromosome 12 open reading frame 61 0.382
C16orf54 chromosome 16 open reading frame 54 0.020
C17orf77 chromosome 17 open reading frame 77 0.394
C19orf66 chromosome 19 open reading frame 66 0.431
C19orf73 chromosome 19 open reading frame 73 0.436
C1orf114 chromosome 1 open reading frame 114 0.031
C1orf194 chromosome 1 open reading frame 194 0.006
C20orf118 chromosome 20 open reading frame 118 0.219
C22orf43 chromosome 22 open reading frame 43 0.410
C2orf57 chromosome 2 open reading frame 57 0.009
C3orf27 chromosome 3 open reading frame 27 0.017
C3orf43 chromosome 3 open reading frame 43 0.019
C5orf64 chromosome 5 open reading frame 64 0.312
C7orf45 chromosome 7 open reading frame 45 0.006
C8orf46 chromosome 8 open reading frame 46 0.040

C9orf152 chromosome 9 open reading frame 152 0.035
C9orf66 chromosome 9 open reading frame 66 0.065
CAPS calcyphosine 0.469
CAPZB capping protein (actin filament) muscle Z‐line, beta 0.465
CCBL1 cysteine conjugate‐beta lyase, cytoplasmic 0.424
CCDC116 coiled‐coil domain containing 116 0.382
CCDC42B coiled‐coil domain containing 42B 0.443
CCL17 chemokine (C‐C motif) ligand 17 0.005
CCR3 chemokine (C‐C motif) receptor 3 0.004
CCR4 chemokine (C‐C motif) receptor 4 0.006
CD14 CD14 molecule 0.051
CD180 CD180 molecule 0.052
CD38 CD38 molecule 0.006
CD7 CD7 molecule 0.234
CDC45 CDC45 cell division cycle 45‐like (S. cerevisiae) 0.269
CDH22 cadherin‐like 22 0.085
CDKL4 cyclin‐dependent kinase‐like 4 0.008
CELF6 bruno‐like 6, RNA binding protein (Drosophila) 0.046
CFH complement factor H 0.042
CFI complement factor I 0.325
CFP complement factor properdin 0.032
CGREF1 cell growth regulator with EF‐hand domain 1 0.457
CHADL chondroadherin‐like 0.441
CHGA chromogranin A (parathyroid secretory protein 1) 0.009
CHRNB2 cholinergic receptor, nicotinic, beta 2 (neuronal) 0.416
CLCN2 chloride channel 2 0.062
CLEC3B C‐type lectin domain family 3, member B 0.418
CNGA1 cyclic nucleotide gated channel alpha 1 0.055
COL11A2 collagen, type XI, alpha 2 0.044
CORO7‐PAM16 CORO7‐PAM16 Read‐Through Transcript 0.094
CORT cortistatin; apoptosis‐inducing, TAF9‐like domain 1 0.331
CTRC chymotrypsin C (caldecrin) 0.024
CXCL11 chemokine (C‐X‐C motif) ligand 11 0.071
CXCL17 chemokine (C‐X‐C motif) ligand 17 0.013
CXCR6 chemokine (C‐X‐C motif) receptor 6 0.241
CXorf56 chromosome X open reading frame 56 0.057
CYBB cytochrome b‐245, beta polypeptide 0.416
CYP11A1 cytochrome P450, family 11, subfamily A, polypeptide 1 0.397
CYP2A6 cytochrome P450, family 2, subfamily A, polypeptide 6 0.013
DCLK1 doublecortin‐like kinase 1 0.357
DENND2D DENN/MADD domain containing 2D 0.038
DES desmin 0.228
DLK1 delta‐like 1 homolog (Drosophila) 0.009
DNAH3 dynein, axonemal, heavy chain 3 0.120

DNAJB8 DnaJ (Hsp40) homolog, subfamily B, member 8 0.012
DOK1 docking protein 1‐like protein; docking protein 1, 62kDa
(downstream of tyrosine kinase 1) 0.388
DPEP2 dipeptidase 2 0.005
DPPA2 developmental pluripotency associated 2 0.011
DPT dermatopontin 0.035
DPYSL5 dihydropyrimidinase‐like 5 0.054
DRP2 dystrophin related protein 2 0.067
DTX3L deltex 3‐like (Drosophila) 0.391
DYDC1 DPY30 domain containing 1 0.039
EFCAB5 EF‐hand calcium binding domain 5 0.038
EFTUD2 elongation factor Tu GTP binding domain containing 2 0.449
EIF2AK2 eukaryotic translation initiation factor 2‐alpha kinase 2 0.433
EIF3CL eukaryotic translation initiation factor 3, subunit C‐like 0.076
EIF4A1
similar to eukaryotic translation initiation factor 4A; small nucleolar
RNA, H/ACA box 67; eukaryotic translation initiation factor 4A,
isoform 1
0.352
ELAC2 elaC homolog 2 (E. coli) 0.383
EPB41L3 erythrocyte membrane protein band 4.1‐like 3 0.052
EPHA4 EPH receptor A4 0.055
ERAP2 endoplasmic reticulum aminopeptidase 2 0.080
ERN2 endoplasmic reticulum to nucleus signaling 2 0.019
F2RL3 coagulation factor II (thrombin) receptor‐like 3 0.360
FABP4 fatty acid binding protein 4, adipocyte 0.080
FAM129C family with sequence similarity 129, member C 0.028
FAM157A family with sequence similarity 157, member A 0.042
FAM162B family with sequence similarity 162, member B 0.006
FAM166B family with sequence similarity 166, member B 0.043
FAM178B family with sequence similarity 178, member B 0.051
FAM46C family with sequence similarity 46, member C 0.414
FAM53A family with sequence similarity 53, member A 0.085
FAM65C family with sequence similarity 65, member C 0.360
FCER1G Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide 0.312
FCGRT Fc fragment of IgG, receptor, transporter, alpha 0.059
FIGF c‐fos induced growth factor (vascular endothelial growth factor D) 0.028
FOXR1 forkhead box R1 0.009
FRMD5 FERM domain containing 5 0.341
FRMD7 FERM domain containing 7 0.012
FZD10 frizzled homolog 10 (Drosophila) 0.385
FZD9 frizzled homolog 9 (Drosophila) 0.422
GAB3 GRB2‐associated binding protein 3 0.069
GABRR2 gamma‐aminobutyric acid (GABA) receptor, rho 2 0.031
GBP5 guanylate binding protein 5 0.068
GCNT7 glucosaminyl (N‐acetyl) transferase family member 7 0.367

GFAP glial fibrillary acidic protein 0.087
GFOD1 glucose‐fructose oxidoreductase domain containing 1 0.499
GIPC2 GIPC PDZ domain containing family, member 2 0.382
GLRB glycine receptor, beta 0.089
GMPR guanosine monophosphate reductase 0.136
GOLGA6A
golgi autoantigen, golgin subfamily a, 6C; golgi autoantigen, golgin
subfamily a, 6 pseudogene; golgi autoantigen, golgin subfamily a, 6D;
golgi autoantigen, golgin subfamily a, 6
0.061
GOLT1A golgi transport 1 homolog A (S. cerevisiae) 0.063
GPN1 GPN‐loop GTPase 1 0.069
GPR20 G protein‐coupled receptor 20 0.208
GPR37 G protein‐coupled receptor 37 (endothelin receptor type B‐like) 0.042
GPR84 G protein‐coupled receptor 84 0.005
GPR97 G protein‐coupled receptor 97 0.421
GPRIN2 G protein regulated inducer of neurite outgrowth 2 0.007
GRAP2 GRB2‐related adaptor protein 2 0.156
HBE1 hemoglobin, epsilon 1 0.362
HBQ1 hemoglobin, theta 1 0.022
HERC6 hect domain and RLD 6 0.226
HES5 hairy and enhancer of split 5 (Drosophila) 0.025
HIPK4 homeodomain interacting protein kinase 4 0.022
HIST1H1A histone cluster 1, H1a 0.031
HIST1H2AH
histone cluster 1, H2ag; histone cluster 1, H2ah; histone cluster 1,
H2ai; histone cluster 1, H2ak; histone cluster 1, H2al; histone cluster
1, H2am
0.449
HIST1H2BL histone cluster 1, H2bl 0.173
HIST4H4
histone cluster 1, H4l; histone cluster 1, H4k; histone cluster 4, H4;
histone cluster 1, H4h; histone cluster 1, H4j; histone cluster 1, H4i;
histone cluster 1, H4d; histone cluster 1, H4c; histone cluster 1, H4f;
histone cluster 1, H4e; histone cluster 1, H4b; histone cluster 1, H4a;
histone cluster 2, H4a; histone cluster 2, H4b
0.376
HMP19 HMP19 protein 0.084
HOXD11 homeobox D11 0.042
HOXD13 homeobox D13 0.197
HPGD hydroxyprostaglandin dehydrogenase 15‐(NAD) 0.196
HPN hepsin 0.023
HRASLS2 HRAS‐like suppressor 2 0.021
HSH2D hematopoietic SH2 domain containing 0.210
HSPB9 heat shock protein, alpha‐crystallin‐related, B9 0.016
HYAL4 hyaluronoglucosaminidase 4 0.013
IFI35 interferon‐induced protein 35 0.371
IFIH1 interferon induced with helicase C domain 1 0.331
IFIT1 interferon‐induced protein with tetratricopeptide repeats 1 0.160
IFIT2 interferon‐induced protein with tetratricopeptide repeats 2 0.215

IFIT5 interferon‐induced protein with tetratricopeptide repeats 5 0.465
IFITM1 interferon induced transmembrane protein 1 (9‐27) 0.143
IFITM3 interferon induced transmembrane protein 3 (1‐8U) 0.459
IFNW1 interferon, omega 1 0.007
IGJ immunoglobulin J polypeptide, linker protein for immunoglobulin
alpha and mu polypeptides 0.008
IGSF6 immunoglobulin superfamily, member 6 0.036
IL13RA2 interleukin 13 receptor, alpha 2 0.384
IL17RE interleukin 17 receptor E 0.069
IL34 interleukin 34 0.496
IL6 interleukin 6 (interferon, beta 2) 0.343
IRF5 interferon regulatory factor 5 0.066
IRGC immunity‐related GTPase family, cinema 0.312
ITIH5 inter‐alpha (globulin) inhibitor H5 0.082
ITK IL2‐inducible T‐cell kinase 0.017
KCNK12 potassium channel, subfamily K, member 12 0.037
KCNRG potassium channel regulator 0.055
KIAA0232 KIAA0232 0.441
KIAA1107 KIAA1107 0.407
KIF5A kinesin family member 5A 0.177
KIRREL2 kin of IRRE like 2 (Drosophila) 0.007
KLRC4 killer cell lectin‐like receptor subfamily C, member 4 0.034
KLRC4‐KLRK1 KLRC4‐KLRK1 Read‐Through Transcript 0.042
KLRD1 killer cell lectin‐like receptor subfamily D, member 1 0.103
KRT80 keratin 80 0.429
LAMP3 lysosomal‐associated membrane protein 3 0.298
LAP3 leucine aminopeptidase 3 0.432
LAT linker for activation of T cells 0.478
LCLAT1 lysocardiolipin acyltransferase 1 0.454
LCN10 lipocalin 10 0.006
LDB3 LIM domain binding 3 0.101
LGALS2 lectin, galactoside‐binding, soluble, 2 0.483
LMAN2L lectin, mannose‐binding 2‐like 0.047
LOC100505676 0.038
LOC100507003 0.059
LOC729020 rcRPE; ribulose‐5‐phosphate‐3‐epimerase 0.057
LPHN1 latrophilin 1 0.447
LPHN3 latrophilin 3 0.033
LRP1B low density lipoprotein‐related protein 1B (deleted in tumors) 0.089
LRRC24 leucine rich repeat containing 24 0.072
LRRD1 Leucine‐rich repeat and death domain‐containing protein 1 0.012
MAK male germ cell‐associated kinase 0.116
MAP1LC3A microtubule‐associated protein 1 light chain 3 alpha 0.044
MARK2 MAP/microtubule affinity‐regulating kinase 2 0.486

MDFI MyoD family inhibitor 0.006
MDK midkine (neurite growth‐promoting factor 2) 0.442
MEFV Mediterranean fever 0.043
MIP major intrinsic protein of lens fiber 0.079
MMRN1 multimerin 1 0.027
MSH4 mutS homolog 4 (E. coli) 0.025
MT1F metallothionein 1F 0.014
MTRNR2L7 MTRNR2‐Like 7 0.059
MUC2 mucin 2, oligomeric mucus/gel‐forming 0.139
MUC5B mucin 5B, oligomeric mucus/gel‐forming 0.105
MX1 myxovirus (influenza virus) resistance 1, interferon‐inducible protein
p78 (mouse) 0.461
MXRA8 matrix‐remodelling associated 8 0.485
MYBPH myosin binding protein H 0.014
MYBPHL myosin binding protein H‐like 0.010
MYO9B myosin IXB 0.342
NAALAD2 N‐acetylated alpha‐linked acidic dipeptidase 2 0.254
NCCRP1 non‐specific cytotoxic cell receptor protein 1 homolog (zebrafish) 0.048
NCF2 neutrophil cytosolic factor 2 0.083
NIPAL2 NIPA‐like domain containing 2 0.204
NMB neuromedin B 0.036
NME5 non‐metastatic cells 5, protein expressed in (nucleoside‐diphosphate
kinase) 0.009
NPFF neuropeptide FF‐amide peptide precursor 0.406
NR0B2 nuclear receptor subfamily 0, group B, member 2 0.010
NR1D2 nuclear receptor subfamily 1, group D, member 2 0.077
NR6A1 nuclear receptor subfamily 6, group A, member 1 0.394
NT5C1B‐RDH14 NT5C1B‐RDH14 Read‐Through Transcript 0.092
OAS1 2',5'‐oligoadenylate synthetase 1, 40/46kDa 0.319
OAS3 2'‐5'‐oligoadenylate synthetase 3, 100kDa 0.264
OR51B4 olfactory receptor, family 51, subfamily B, member 4 0.005
OR51B5 olfactory receptor, family 51, subfamily B, member 5 0.024
OR52N4 olfactory receptor, family 52, subfamily N, member 4 0.006
OR7E24 olfactory receptor, family 7, subfamily E, member 24 0.054
OTX1 orthodenticle homeobox 1 0.430
P2RY4 pyrimidinergic receptor P2Y, G‐protein coupled, 4 0.009
PABPC4L poly(A) binding protein, cytoplasmic 4‐like 0.005
PADI4 peptidyl arginine deiminase, type IV 0.016
PARD3B par‐3 partitioning defective 3 homolog B (C. elegans) 0.091
PARP10 poly (ADP‐ribose) polymerase family, member 10 0.492
PARP12 poly (ADP‐ribose) polymerase family, member 12 0.412
PARP14 poly (ADP‐ribose) polymerase family, member 14 0.432
PARP9 poly (ADP‐ribose) polymerase family, member 9 0.317
PCDHB16 protocadherin beta 16 0.209

PCDHB6 protocadherin beta 6 0.050
PCDHB8 protocadherin beta 8 0.374
PCDHGA1 protocadherin gamma subfamily A, 1 0.036
PCDHGA4 protocadherin gamma subfamily A, 4 0.034
PCDHGA5 protocadherin gamma subfamily A, 5 0.040
PCDHGA6 protocadherin gamma subfamily A, 6 0.041
PCDHGB2 protocadherin gamma subfamily B, 2 0.052
PCDHGB7 protocadherin gamma subfamily B, 7 0.365
PCSK5 proprotein convertase subtilisin/kexin type 5 0.059
PCSK6 proprotein convertase subtilisin/kexin type 6 0.496
PDHB pyruvate dehydrogenase (lipoamide) beta 0.491
PELI3 pellino homolog 3 (Drosophila) 0.485
PGBD5 piggyBac transposable element derived 5 0.416
PGPEP1L pyroglutamyl‐peptidase 1‐like 0.042
PIK3AP1 phosphoinositide‐3‐kinase adaptor protein 1 0.394
PLSCR1 phospholipid scramblase 1 0.362
PLVAP plasmalemma vesicle associated protein 0.029
PMF1‐BGLAP PMF1‐BGLAP Read‐Through Transcript 0.457
PMFBP1 polyamine modulated factor 1 binding protein 1 0.408
PNPT1 polyribonucleotide nucleotidyltransferase 1 0.420
POC1B‐GALNT4 POC1B‐GALNT4 Read‐Through Transcript 0.122
POF1B premature ovarian failure, 1B 0.021
PPARD peroxisome proliferator‐activated receptor delta 0.411
PPP6C protein phosphatase 6, catalytic subunit 0.498
PRELP proline/arginine‐rich end leucine‐rich repeat protein 0.092
PRICKLE1 prickle homolog 1 (Drosophila) 0.056
PROX2 prospero homeobox 2 0.093
PRPH2 peripherin 2 (retinal degeneration, slow) 0.016
PRR24 hypothetical protein LOC255783 0.272
PRR5‐ARHGAP8 Rho GTPase activating protein 8; proline rich 5 (renal);
PRR5‐ARHGAP8 fusion 0.051
PRRG2 proline rich Gla (G‐carboxyglutamic acid) 2 0.278
PSMD1 proteasome (prosome, macropain) 26S subunit, non‐ATPase, 1 0.061
PSTPIP1 proline‐serine‐threonine phosphatase interacting protein 1 0.014
PTGDS prostaglandin D2 synthase, hematopoietic; prostaglandin D2
synthase 21kDa (brain) 0.035
PTPRB protein tyrosine phosphatase, receptor type, B 0.129
PTX4 chromosome 16 open reading frame 38 0.005
RAB17 RAB17, member RAS oncogene family 0.025
RAB9B RAB9B, member RAS oncogene family 0.020
RAET1G retinoic acid early transcript 1G 0.410
RAG2 recombination activating gene 2 0.020
RAP1GDS1 RAP1, GTP‐GDP dissociation stimulator 1 0.497
RARRES3 retinoic acid receptor responder (tazarotene induced) 3 0.299

RBP1 retinol binding protein 1, cellular 0.360
RIMBP3B RIMS binding protein 3B; RIMS binding protein 3C; RIMS binding
protein 3 0.132
RIMBP3C RIMS binding protein 3B; RIMS binding protein 3C; RIMS binding
protein 3 0.328
RNF183 ring finger protein 183 0.006631
RPRM reprimo, TP53 dependent G2 arrest mediator candidate 0.008019
RSAD2 radical S‐adenosyl methionine domain containing 2 0.196522
RUSC1‐AS1 RUSC1 antisense RNA 1 0.497053
SAMD5 sterile alpha motif domain containing 5 0.127803
SAMHD1 SAM domain and HD domain 1 0.328355
SAP25 Sin3A‐associated protein 0.397751
SCRT1 scratch homolog 1, zinc finger protein (Drosophila) 0.02862
SDHD
similar to succinate dehydrogenase complex, subunit D, integral
membrane protein; succinate dehydrogenase complex, subunit D,
integral membrane protein
0.060534
SECTM1 secreted and transmembrane 1 0.431267
SEMA6A sema domain, transmembrane domain (TM), and cytoplasmic
domain, (semaphorin) 6A 0.488318
SERPINB10 serpin peptidase inhibitor, clade B (ovalbumin), member 10 0.017739
SERPINB2 serpin peptidase inhibitor, clade B (ovalbumin), member 2 0.460325
SFI1 Sfi1 homolog, spindle assembly associated (yeast) 0.453148
SGPP2 sphingosine‐1‐phosphate phosphotase 2 0.034786
SHBG sex hormone‐binding globulin 0.052575
SHMT2 serine hydroxymethyltransferase 2 (mitochondrial) 0.042054
SIDT1 SID1 transmembrane family, member 1 0.009828
SLC10A1 solute carrier family 10 (sodium/bile acid cotransporter family),
member 1 0.007138
SLC25A13 solute carrier family 25, member 13 (citrin) 0.300565
SLC25A41 solute carrier family 25, member 41 0.309854
SLC46A2 solute carrier family 46, member 2 0.03935
SLC47A2 solute carrier family 47, member 2 0.02182
SLC7A7 solute carrier family 7 (cationic amino acid transporter, y+ system),
member 7 0.088014
SLC7A8 solute carrier family 7 (cationic amino acid transporter, y+ system),
member 8 0.075226
SLCO1C1 solute carrier organic anion transporter family, member 1C1 0.040872
SLFN11 schlafen family member 11 0.251936
SLFNL1 schlafen‐like 1 0.434731
SMPDL3B sphingomyelin phosphodiesterase, acid‐like 3B 0.011364
SP110 SP110 nuclear body protein 0.044318
SPRED3 sprouty‐related, EVH1 domain containing 3 0.041259
SRL sarcalumenin 0.480133
ST18 suppression of tumorigenicity 18 (breast carcinoma) (zinc finger 0.01131

protein)
ST3GAL5 ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 5 0.436894
ST3GAL6 ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 6 0.058884
ST8SIA5 ST8 alpha‐N‐acetyl‐neuraminide alpha‐2,8‐sialyltransferase 5 0.040193
STOML1 stomatin (EPB72)‐like 1 0.042074
STOX1 storkhead box 1 0.479429
STX19 syntaxin 19 0.060201
STXBP2 syntaxin binding protein 2 0.43449
STXBP5L8 syntaxin binding protein 5‐like 0.169636
SYNJ2BP‐COX16 SYNJ2BP‐COX16 Read‐Through Transcript 0.038542
SYNPO2 synaptopodin 2 0.450861
TAL2 T‐cell acute lymphocytic leukemia 2 0.024599
TAS2R30 taste receptor, type 2, member 30 0.034361
TBC1D28 TBC1 domain family, member 28 0.014994
TBC1D29 TBC1 domain family, member 29 0.007344
TBRG4 transforming growth factor beta regulator 4 0.464622
TCTE1 t‐complex‐associated‐testis‐expressed 1 0.009037
TEX19 testis expressed 19 0.482793
TGIF2‐C20ORF24 TGIF2‐C20ORF24 Read‐Through Transcript 0.032766
THTPA thiamine triphosphatase 0.46625
TLR3 toll‐like receptor 3 0.39754
TMC6 transmembrane channel‐like 6 0.468143
TMEM14E transmembrane protein 14E 0.006931
TMEM183B transmembrane protein 183A; transmembrane protein 183B 0.392782
TMEM229B chromosome 14 open reading frame 83 0.455182
TMEM232 hypothetical protein LOC642987 0.474396
TMEM51 transmembrane protein 51 0.039565
TMPRSS6 transmembrane protease, serine 6 0.306026
TNFAIP8L2 tumor necrosis factor, alpha‐induced protein 8‐like 2 0.019201
TNFRSF8 tumor necrosis factor receptor superfamily, member 8 0.054544
TNFSF10 tumor necrosis factor (ligand) superfamily, member 10 0.467522
TNNI3 troponin I type 3 (cardiac) 0.017875
TOMM40 translocase of outer mitochondrial membrane 40 homolog (yeast) 0.407491
TPTE transmembrane phosphatase with tensin homology 0.065192
TRAF3IP1 TNF receptor‐associated factor 3 interacting protein 1 0.47951
TRANK1 lupus brain antigen 1 0.238881
TRIM14 tripartite motif‐containing 14 0.408594
TRIM2 tripartite motif‐containing 2 0.474502
TRIM21 tripartite motif‐containing 21 0.40533
TRIM34 TRIM6‐TRIM34 readthrough transcript; tripartite motif‐containing 6;
tripartite motif‐containing 34 0.341296
TRIM60 tripartite motif‐containing 60 0.014608
TRIML1 tripartite motif family‐like 1 0.381682
TSN translin 0.461096

TTC23L tetratricopeptide repeat domain 23‐like 0.007338
TUBA3D tubulin, alpha 3d; tubulin, alpha 3c 0.379318
U2AF1L4 U2 small nuclear RNA auxiliary factor 1‐like 4 0.473631
UBA7 ubiquitin‐like modifier activating enzyme 7 0.261071
ULK2 unc‐51‐like kinase 2 (C. elegans) 0.289878
UNC5D unc‐5 homolog D (C. elegans) 0.039076
USH1G Usher syndrome 1G (autosomal recessive) 0.019104
USP18 ubiquitin specific peptidase 18 0.317838
UXT ubiquitously‐expressed transcript 0.439903
VMO1 vitelline membrane outer layer 1 homolog (chicken) 0.01623
VTN vitronectin 0.21341
WDR86 WD repeat domain 86 0.380295
WDR87 WD repeat domain 87 0.067823
WDR93 WD repeat domain 93 0.077552
WNT2B wingless‐type MMTV integration site family, member 2B 0.032417
XYLT1 xylosyltransferase I 0.113656
YTHDF2 YTH domain family, member 2 0.470145
ZBTB32 zinc finger and BTB domain containing 32 0.44698
ZC4H2 zinc finger, C4H2 domain containing 0.055356
ZFAT zinc finger and AT hook domain containing 0.444769
ZKSCAN5 zinc finger with KRAB and SCAN domains 5 0.447756
ZNF142 zinc finger protein 142 0.464554
ZNF268 zinc finger protein 268 0.047543
ZNF32 zinc finger protein 32 0.018522
ZNF439 zinc finger protein 439 0.200902
ZNF512 zinc finger protein 512 0.493183
ZNF560 zinc finger protein 560 0.015019
ZNF562 zinc finger protein 562 0.141053
ZNF716 zinc finger protein 716 0.333159
ZNF80 zinc finger protein 80 0.460032
ZNF835 zinc finger protein 835 0.029205
ZNF878 similar to zinc finger protein 709 0.03129
ZPLD1 zona pellucida‐like domain containing 1 0.027586

Supplementary Table 6. GO analysis of different expression genes after PBX3‐overexpression by DAVID database via
biological process terms
Term Count % PValue Genes
Cell surface
receptor linked
signal
transduction
98 11.34 3.2E‐02
F2RL3, ADCY2, OR4F21, GRIK2, LHCGR, CXCL11, MARCO,
LPHN3, APOA1, GAB2, ELTD1, PDGFC, CIB2, ADAM8, ROS1,
KLRD1, LPHN1, WNT10A, FGF22, GEM, THY1, IGSF6,
GABRR2, CCR4, CCR3, ITGAL, IFITM1, HMP19, ITGBL1,
ADAP1, OR52N4, P2RY4, TEK, OR51B5, ZAP70, FCER1G,
OR51B4, KLRF1, GPR20, ANGPTL3, PLAT, GPR158,
TNFRSF13B, KCNK2, NPY5R, NOTCH3, SEMA6A, GNGT1,
EPHA4, LAT, EMR1, GPR37, NPW, OR6K3, CD79A, AREG,
CORT, LHB, ADRA1D, GPR84, GPR85, TAS2R5, LEPR, TACR1,
GNRHR, GABBR2, OR7E24, SCT, CXCR5, OR13A1, CXCR6,
NOS3, APLN, NPFF, GPR97, GPR176, DOK1, PTAFR, NKD2,
FFAR1, ABI1, NMB, SCG5, FIGF, CD7, FZD9, COL4A3, PTPRC,
GLRB, PTPRE, DGKI, CISH, CCL17, WNT2B, FZD10, TAS2R30,
HPGD, CD14
Immune response 63 7.29 4.5E‐08
TLR3, BNIP3, VTN, CXCL11, TLR9, CFP, RAET1G, IL1RAP,
TICAM2, CFH, ERAP2, CFI, APLN, LAIR1, GBP5, BST2, NCF2,
BST1, STXBP2, YTHDF2, GEM, CNGA1, SIGIRR, CTSW, IGSF6,
CCR4, TREM1, PTAFR, TNFAIP8L2, ITGAL, C7, IFIH1, IFITM3,
OAS3, RAG1, RSAD2, OAS1, FCGRT, CD70, RAG2, IFI35,
FCGR1A, ZAP70, HLA‐DRB4, FCER1G, C2, CD7, SECTM1,
PTPRC, IL6, TNFRSF13B, IGJ, SAMHD1, LOC554223, AIM2,
CD180, CCL17, LAT, TNFSF10, CYBB, AIRE, CD79A, CD14
Cell adhesion 54 6.25 6.3E‐05
CLDN7, PPARD, PCDHA9, MYBPC2, MYBPC3, PCDHGA6, VTN,
PCDHGA5, PCDHGA4, MMRN1, PCDHGA1, CDH22,
CNTNAP2, COL11A2, KIRREL2, ADAM8, BOC, NEGR1,
SPACA4, CLCA2, PCDHB8, PCDHB6, PCDHGB7, CPXM2,
PCDHGB2, BCL2L11, THY1, AMBP, HES5, CCR3, ITGAL,
PPFIA1, PKD1L1, SCARF2, ITGBL1, SEMA5A, LY6D, PCDHB16,
MYBPH, TEK, PSTPIP1, ANGPTL3, GPNMB, DPT, HAPLN2,
COL4A3, PTPRC, LGALS4, COL13A1, PRPH2, OMD, EMR1,
OMG, MUC5B
Ion transport 46 5.32 2.7E‐02
SLC5A2, KCNC1, CLCN2, GRIK2, SLC22A14, SLC22A11, KCNRG,
FKBP1B, KCNK12, KCNJ14, BEST4, MARCO, BEST2, KCNQ5,
KCNS1, SLCO1A2, P2RY4, ANO2, SLCO1C1, KCNE3, PTPRC,
CACNA2D1, CLCA2, GLRB, KCND2, SLC8A2, SCN2B, KCNB1,
AMACR, TOMM40, KCNK2, CNGA1, SLC10A1, GABRR2, CYBB,
SLC25A13, CLIC3, SLC6A8, P2RX3, ATP2A1, SLCO1B1,
CHRNB4, CHRNB2, ATP6V0A4, CHRNG, KCTD12
Cell‐cell signaling 45 5.21 5.2E‐04
DHH, CPLX3, GRIK2, SOX2, FFAR1, CD70, NMB, GABBR2,
CXCL11, FKBP1B, HOXD11, SEMA5A, KCNQ5, PCDHB16, TEK,
SYN2, PCSK5, NPFF, PLAT, STX1A, IL6, GLRB, GPR176, KCND2,

BST2, SCN2B, KIF5A, PCDHB6, PARK2, NPY5R, CCL17,
GABRR2, NAALAD2, ATXN3, TNFSF10, CAMK4, P2RX3,
CHRNB4, CHRNB2, AREG, GRAP2, CORT, LHB, ADRA1D,
NR5A1
Homeostatic
process 45 5.21 2.9E‐02
DHH, C7, PPARD, GRIK2, PDIA2, TACR1, FGGY, PRND, FFAR1,
SLC7A8, BNIP3, FKBP1B, ACSBG1, BEST2, APOA1, P2RY4,
HAAO, COL11A2, ANGPTL3, APOM, GAL3ST1, MB, CD7,
MUC2, PTPRC, IL6, GLRB, KCND2, TNFRSF13B, MTL5, TNNI3,
BCL2L11, GIMAP1, CD38, HIF1A, CCR4, CCR3, ATP2A1,
CHRNB4, FABP4, CHRNB2, LIPC, ATP6V0A4, NR5A2, CHRNG
Inflammatory
response 28 3.24 1.1E‐03
C7, ITGAL, TACR1, TLR3, CXCL11, TLR9, CFP, IL17D, MEFV,
IL1RAP, TICAM2, CFH, C2, CFI, IL6, CD180, CCL17, SIGIRR,
APOL3, CYBB, HIF1A, SERPINF2, CCR4, CCR3, CARD18,
ALOX5, CD14, PTAFR
Regulation of
system process 26 3.01 2.3E‐03
CPLX3, GRIK2, TACR1, BTC, MYBPC3, RPS6KB1, FKBP1B,
APOA1, DES, MYBPH, NOS3, PLAT, STX1A, NTF4, PARK2,
TNNI3, NPY5R, CD38, P2RX3, HSPB7, ATP2A1, CHRNB4,
CHRNB2, ALOX5, PBX3, ADRA1D
Regulation of
hydrolase activity 22 2.55 6.5E‐02
FGD2, TBC1D3C, COL4A3, F2RL3, LEPR, TACR1, MYBPC3,
LHCGR, RAG1, TNNI3, FKBP1B, ADAP1, THY1, APOA1, ADAP2,
TBC1D29, P2RY4, TBC1D28, NOS3, CSTA, ANGPTL3, FGD5
Steroid metabolic
process 18 2.08 7.4E‐03
PPARD, CYP11A1, LEPR, PRKAG2, AMACR, DHRS9, NR0B2,
UGT2B17, CYP39A1, APOA1, AKR1B15, APOF, SULT1A2,
UGT2B15, NR5A2, ANGPTL3, LIPC, LHB
Fatty acid
metabolic process 16 1.85 2.7E‐02
PPARD, ALOXE3, ACSM2A, PRKAG2, ACSBG1, ACSM3,
PRKAR2B, PTGDS, ALOX12B, FABP4, ELOVL7, ALOX5, LIPC,
ANGPTL3, HPGD, ACSL6
Regulation of
membrane
potential
14 1.62 5.8E‐03DHH, GLRB, PPARD, KCND2, GRIK2, TACR1, BNIP3, GIMAP1,
ACSBG1, BEST2, CHRNB4, CHRNB2, GAL3ST1, CHRNG
Response to
hypoxia 12 1.39 3.3E‐02
PLAT, CD38, NOL3, HIF1A, CA9, PDIA2, P2RX3, ALDOC, BNIP3,
NOS3, CHRNB2, MB

Supplementary Table 7. Embryo development associated genes up‐regulated by PBX3 DAVID ID Gene name RatioA1BG alpha-1-B glycoprotein 41.51 ABHD1 abhydrolase domain containing 1 2.83 ACOT6 acyl-CoA thioesterase 6 2.64 ACSL6 acyl-CoA synthetase long-chain family member 6 8.24 ACSM2A acyl-CoA synthetase medium-chain family member 2A 67.83 ADCY2 adenylate cyclase 2 (brain) 30.73 AHSP erythroid associated factor 476.19 ALOX5 arachidonate 5-lipoxygenase 16.66 ALOXE3 arachidonate lipoxygenase 3 28.09 AMBP alpha-1-microglobulin/bikunin precursor 2.19 AMN amnionless homolog (mouse) 32.11 ANGPTL3 angiopoietin-like 3 254.27 ANK2 ankyrin 2, neuronal 16.88 ANKRD45 ankyrin repeat domain 45 19.49 APCDD1L adenomatosis polyposis coli down-regulated 1-like 29.48 APLN apelin 2.47 APOA1 apolipoprotein A-I 41.18
APOBEC2 apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
40.57
ARHGAP15
Rho GTPase activating protein 15 19.52
ARHGAP19
Rho GTPase activating protein 19 11.40
ASGR1 asialoglycoprotein receptor 1 37.87 ASPN asporin 79.26 ATP6V0A4
ATPase, H+ transporting, lysosomal V0 subunit a4 29.29
BCL2L14 BCL2-like 14 (apoptosis facilitator) 98.77 BCL2L2 BCL2-like 2 2.14 BEST2 bestrophin 2 148.04 BEST4 bestrophin 4 2.90 BMP5 bone morphogenetic protein 5 10.17 BOC Boc homolog (mouse) 17.75 C11orf16 chromosome 11 open reading frame 16 18.00 C1orf228 chromosome 1 open reading frame 228 2.04 C2orf72 chromosome 2 open reading frame 72 34.96 C8orf74 chromosome 8 open reading frame 74 294.85 C9orf50 chromosome 9 open reading frame 50 151.89 CACNA2D1
calcium channel, voltage-dependent, alpha 2/delta subunit 1 2884.7
9 CAMK4 calcium/calmodulin-dependent protein kinase IV 11.77 CAPN5 calpain 5 4.75

CARD18 caspase recruitment domain family, member 18 14.45 CASC1 cancer susceptibility candidate 1 26.48 CCDC122 coiled-coil domain containing 122 2.66 CCDC149 coiled-coil domain containing 149 3.37 CCDC17 coiled-coil domain containing 17 15.76 CCDC33 coiled-coil domain containing 33 37.22 CCDC62 coiled-coil domain containing 62 2.58 CEACAM19
carcinoembryonic antigen-related cell adhesion molecule 19 15.11
CHD5 chromodomain helicase DNA binding protein 5 2.26 CHRNG cholinergic receptor, nicotinic, gamma 32.30 CIB2 calcium and integrin binding family member 2 27.21 CLGN calmegin 2.11 CLIC3 chloride intracellular channel 3 2.52 CLIP3 CAP-GLY domain containing linker protein 3 2.81 CNIH3 cornichon homolog 3 (Drosophila) 19.49 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) 4.81 CPLX3 complexin 3 71.78 CRYGS crystallin, gamma S 47.87 CYP39A1 cytochrome P450, family 39, subfamily A, polypeptide 1 2.86 CYP8B1 cytochrome P450, family 8, subfamily B, polypeptide 1 50.99 DBNDD1 dysbindin (dystrobrevin binding protein 1) domain containing 1 17.44 DHH desert hedgehog homolog (Drosophila) 27.78 DIRAS1 DIRAS family, GTP-binding RAS-like 1 7.16 DNAJB5 DnaJ (Hsp40) homolog, subfamily B, member 5 2.25 DNAJC28 DnaJ (Hsp40) homolog, subfamily C, member 28 2.23 DNALI1 dynein, axonemal, light intermediate chain 1 22.53 DPPA4 developmental pluripotency associated 4 11.40 DSE dermatan sulfate epimerase 2.70 EGFL8 EGF-like-domain, multiple 8 2.44 EIF4ENIF1
eukaryotic translation initiation factor 4E nuclear import factor 1
2.10
ELAVL3 ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
30.49
EMILIN3 elastin microfibril interfacer 3 2.95
EMR1 egf-like module containing, mucin-like, hormone receptor-like 1
12.87
ENPP7 ectonucleotide pyrophosphatase/phosphodiesterase 7 32.87
ESF1 similar to ABT1-associated protein; ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae)
11.30
FAM109B family with sequence similarity 109, member B 13.31 FAM117A family with sequence similarity 117, member A 3.84 FAM180A family with sequence similarity 180, member A 282.44 FAM19A3 family with sequence similarity 19 (chemokine (C-C 24.31

motif)-like), member A3 FAM25C family with sequence similarity 25, member C 300.60 FAM92B family with sequence similarity 92, member B 106.17 FBLN2 fibulin 2 19.19 FCGR1A Fc fragment of IgG, high affinity Ia, receptor (CD64) 13.19 FER1L5 fer-1-like 5 (C. elegans) 25.03 FGD5 FYVE, RhoGEF and PH domain containing 5 108.00 FGGY FGGY carbohydrate kinase domain containing 32.46 FLG2 filaggrin family member 2 10.52 FLJ22184 hypothetical protein FLJ22184 2.25 FOXD4L1 forkhead box D4-like 1 40.44 FOXS1 forkhead box S1 148.86 FSIP2 fibrous sheath interacting protein 2 13.29 GABBR2 gamma-aminobutyric acid (GABA) B receptor, 2 8.50 GAL3ST1 galactose-3-O-sulfotransferase 1 128.88 GLIS1 GLIS family zinc finger 1 21.91 GNRHR gonadotropin-releasing hormone receptor 9.92 GOLGA6D
golgi autoantigen, golgin subfamily a, 6D 12.41
GPIHBP1 glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1
19.81
GPR176 G protein-coupled receptor 176 12.62 GPR85 G protein-coupled receptor 85 29.09 GRIK2 glutamate receptor, ionotropic, kainate 2 4.20 GRIP2 glutamate receptor interacting protein 2 2.75 GSTA2 glutathione S-transferase alpha 2 152.58 HAAO 3-hydroxyanthranilate 3,4-dioxygenase 154.80 HIC1 hypermethylated in cancer 1 24.58 HIST1H3J histone cluster 1, H3j 2.00 HIST1H4A
histone cluster 1, H4b; histone cluster 1, H4a; 19.39
HOXD3 homeobox D3 2.29 HS3ST3B1 heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 2.05 IL1RAP interleukin 1 receptor accessory protein 33.89 INSM2 insulinoma-associated 2 13.90 IQUB IQ motif and ubiquitin domain containing 3.05 JAZF1 JAZF zinc finger 1 21.97 KATNAL1 katanin p60 subunit A-like 1 2.63
KCNB1 potassium voltage-gated channel, Shab-related subfamily, member 1
131.34
KCNC1 potassium voltage-gated channel, Shaw-related subfamily, member 1
43.60
KCNJ14 potassium inwardly-rectifying channel, subfamily J, member 14 2.12 KCNK2 potassium channel, subfamily K, member 2 19.87

KCNQ5 potassium voltage-gated channel, KQT-like subfamily, member 5
11.18
KIAA1045 KIAA1045 15.08 KIAA1683 KIAA1683 2.34 KIF26B kinesin family member 26B 19.26 KLHL32 kelch-like 32 (Drosophila) 6.06 KLRF1 killer cell lectin-like receptor subfamily F, member 1 40.41 KRT32 keratin 32 426.17 LEPR leptin receptor 15.92 LHB luteinizing hormone beta polypeptide 25.42 LHCGR luteinizing hormone/choriogonadotropin receptor 14.78 LIN28A lin-28 homolog (C. elegans) 10.45 LOC554223
hypothetical LOC554223 86.05
LOC728392
hypothetical protein LOC728392 2.75
LRRC17 leucine rich repeat containing 17 14.84 LRRC39 leucine rich repeat containing 39 20.81
MAF v-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian)
47.09
MAGI2 membrane associated guanylate kinase, WW and PDZ domain containing 2
4.51
MAT1A methionine adenosyltransferase I, alpha 28.12 MED12L mediator complex subunit 12-like 4.51 MFSD2B hypothetical protein LOC388931 163.57 MSI1 musashi homolog 1 (Drosophila) 67.82 MTL5 metallothionein-like 5, testis-specific (tesmin) 3.79 MYBPC2 myosin binding protein C, fast type 39.58 MYBPC3 myosin binding protein C, cardiac 3.24 MYLK3 myosin light chain kinase 3 2.67 MYO1A myosin IA 13.60 MYOZ1 myozenin 1 53.69 NEGR1 neuronal growth regulator 1 3.43 NKD2 naked cuticle homolog 2 (Drosophila) 14.87 NOTCH3 Notch homolog 3 (Drosophila) 17.18 NOVA2 neuro-oncological ventral antigen 2 185.06
NPL N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase)
11.92
NPW neuropeptide W 2.16 NPY5R neuropeptide Y receptor Y5 73.56 NR5A1 nuclear receptor subfamily 5, group A, member 1 14.78 NR5A2 nuclear receptor subfamily 5, group A, member 2 30.12 NTF4 neurotrophin 4 450.80 NYNRIN KIAA1305 38.45

ODAM odontogenic, ameloblast asssociated 55.84 OLFM2 olfactomedin 2 29.83 OMD osteomodulin 112.05 OMG oligodendrocyte myelin glycoprotein 2.07 P2RX3 purinergic receptor P2X, ligand-gated ion channel, 3 60.93 P4HA1 prolyl 4-hydroxylase, alpha polypeptide I 8.17 PALM3 Paralemmin-3 50.43 PAPPA2 pappalysin 2 46.30 PBX3 pre-B-cell leukemia homeobox 3 88.94 PCDHA9 protocadherin alpha 9 2.16 PCOLCE procollagen C-endopeptidase enhancer 211.29 PDIA2 protein disulfide isomerase family A, member 2 233.37 PDLIM4 PDZ and LIM domain 4 19.77 PKD1L1 polycystic kidney disease 1 like 1 14.95 PLA2G4D phospholipase A2, group IVD (cytosolic) 130.30 PLAG1 pleiomorphic adenoma gene 1 18.21 PLXNA4 plexin A4 39.31
PPFIA4 protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4
4.41
PPP1R1C protein phosphatase 1, regulatory (inhibitor) subunit 1C 223.65 PRKAR2B protein kinase, cAMP-dependent, regulatory, type II, beta 35.87 PRND prion protein 2 (dublet) 2.52 PTPRR protein tyrosine phosphatase, receptor type, R 54.15 RAG1 recombination activating gene 1 3.74 RAPGEF5 Rap guanine nucleotide exchange factor (GEF) 5 30.38
RBPJL recombination signal binding protein for immunoglobulin kappa J region-like
27.77
RFESD Rieske (Fe-S) domain containing 2.48 RFX3 regulatory factor X, 3 (influences HLA class II expression) 23.46 RGAG1 retrotransposon gag domain containing 1 3.24 RGMA RGM domain family, member A 18.12 RNF182 ring finger protein 182 2.13 ROS1 c-ros oncogene 1 , receptor tyrosine kinase 2.11 RPS6KB1 ribosomal protein S6 kinase, 70kDa, polypeptide 1 2.21 RTDR1 rhabdoid tumor deletion region gene 1 39.71 SALL2 sal-like 2 (Drosophila) 6.24 SCARF2 scavenger receptor class F, member 2 12.00 SCG5 secretogranin V (7B2 protein) 36.45 SCGB1D2 secretoglobin, family 1D, member 2 627.37 SCN2B sodium channel, voltage-gated, type II, beta 8.43 SDC2 syndecan 2 6.96
SEMA5A sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A
2.44

SERPINC1
serpin peptidase inhibitor, clade C (antithrombin), member 1 31.09
SERPINF2
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2
17.57
SGK2 serum/glucocorticoid regulated kinase 2 20.06 SGOL1 shugoshin-like 1 (S. pombe) 9.40 SHISA7 UPF0626 protein A 10.58 SIAH3 seven in absentia homolog 3 (Drosophila) 35.56
SLC22A11 solute carrier family 22 (organic anion/urate transporter), member 11
111.92
SLC22A14 solute carrier family 22, member 14 36.99 SLC43A1 solute carrier family 43, member 1 44.14 SLC8A2 solute carrier family 8 (sodium/calcium exchanger), member 2 32.53 SLCO1A2 solute carrier organic anion transporter family, member 1A2 7.14 SLCO1B1 solute carrier organic anion transporter family, member 1B1 101.72 SNX20 sorting nexin 20 43.10 SOX2 SRY (sex determining region Y)-box 2 3.74 SPACA4 sperm acrosome associated 4 26.29 SPTY2D1 SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) 24.41
SULT1A2 sulfotransferase family, cytosolic, 1A, phenol-preferring, member 2
2.22
SYCP2 synaptonemal complex protein 2 2.00
TAF7L TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa
16.89
TBC1D3C TBC1 domain family, member 3B; TBC1 domain family, member 3C
22.31
TEK TEK tyrosine kinase, endothelial 25.59 TMEM119 transmembrane protein 119 52.73 TMEM191C
transmembrane protein 191B; transmembrane protein 191C 34.32
TMEM63C
transmembrane protein 63C 2.34
TMEM88 transmembrane protein 88 34.92 TOX thymocyte selection-associated high mobility group box 168.89 TP53TG5 TP53 target 5 18.43 TRIM63 tripartite motif-containing 63 197.36 TRIM71 tripartite motif-containing 71 78.61 TSPAN5 tetraspanin 5 3.51 UBQLNL ubiquilin-like 16.96 UCHL1 ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) 44.44 UGT2B17 UDP glucuronosyltransferase 2 family, polypeptide B17 19.99
WFIKKN1 WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1
12.80
WNT10A wingless-type MMTV integration site family, member 10A 189.62

ZAP70 zeta-chain (TCR) associated protein kinase 70kDa 33.22 ZCCHC12 zinc finger, CCHC domain containing 12 24.72 ZDHHC22 zinc finger, DHHC-type containing 22 41.79 ZFHX4 zinc finger homeobox 4 27.12 ZFR2 zinc finger RNA binding protein 2 43.64 ZNF521 zinc finger protein 521 78.12 ZNF649 zinc finger protein 649 14.46

Supplementary Table 8. Genes directly regulated by PBX3 as revealed by RNA‐seq and ChIP‐seq
analysis
Gene symbol Gene name Ratio
Up‐regulated genes
ABCB1 ATP‐binding cassette, sub‐family B (MDR/TAP), member 1 3.89
ABI1 abl‐interactor 1 2.64
ADCY2 adenylate cyclase 2 (brain) 30.73
ADPGK ADP‐dependent glucokinase 31.65
ANK1 ankyrin 1, erythrocytic 10.05
ANK2 ankyrin 2, neuronal 16.88
ANKRD12 ankyrin repeat domain 12 2.45
ANKRD22 ankyrin repeat domain 22 2.23
ANO2 anoctamin 2 10.38
ARHGAP15 Rho GTPase activating protein 15 19.52
ARHGAP19 Rho GTPase activating protein 19 11.40
ARL17B ADP‐ribosylation factor‐like 17 pseudogene 1; ADP‐ribosylation
factor‐like 17 2.09
ARSD arylsulfatase D 27.50
ART1 ADP‐ribosyltransferase 1 150.86
ASPN asporin 79.26
ATF3 activating transcription factor 3 3.05
ATG9B ATG9 autophagy related 9 homolog B (S. cerevisiae) 2.67
ATP6V0A4 ATPase, H+ transporting, lysosomal V0 subunit a4 29.29
ATXN3 ataxin 3 2.16
BCL2L14 BCL2‐like 14 (apoptosis facilitator) 98.77
BMP5 bone morphogenetic protein 5 10.17
BNIP3 BCL2/adenovirus E1B 19kDa interacting protein 3 3.65
BOC Boc homolog (mouse) 17.75
C10orf10 chromosome 10 open reading frame 10 2.41
C11orf16 chromosome 11 open reading frame 16 18.00
C1orf116 chromosome 1 open reading frame 116 10.60
C1orf21 chromosome 1 open reading frame 21 2.61
C1orf228 chromosome 1 open reading frame 228 2.04
C6orf201 chromosome 6 open reading frame 201 3.74
C7orf65 chromosome 7 open reading frame 65 27.82
C8orf86 chromosome 8 open reading frame 86 167.46
CACNA2D1 calcium channel, voltage‐dependent, alpha 2/delta subunit 1 2884.80
CAMK4 calcium/calmodulin‐dependent protein kinase IV 11.77
CASC1 cancer susceptibility candidate 1 26.48
CCDC106 coiled‐coil domain containing 106 2.46
CCDC114 coiled‐coil domain containing 114 3.61
CCDC149 coiled‐coil domain containing 149 3.37
CCDC33 coiled‐coil domain containing 33 37.22
CCDC62 coiled‐coil domain containing 62 2.58

CD70 CD70 molecule 2.12
CES3 carboxylesterase 3 9.76
CHD5 chromodomain helicase DNA binding protein 5 2.26
CIB2 calcium and integrin binding family member 2 27.21
CLGN calmegin 2.11
CLUL1 clusterin‐like 1 (retinal) 13.16
CNIH3 cornichon homolog 3 (Drosophila) 19.49
CNTNAP2 contactin associated protein‐like 2 3.05
COL13A1 collagen, type XIII, alpha 1 35.02
COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) 4.81
CPNE5 copine V 73.87
CPXM2 carboxypeptidase X (M14 family), member 2 3.41
CYP39A1 cytochrome P450, family 39, subfamily A, polypeptide 1 2.86
DGKI diacylglycerol kinase, iota 9.37
DHRS9 dehydrogenase/reductase (SDR family) member 9 7.25
DIRAS1 DIRAS family, GTP‐binding RAS‐like 1 7.16
DSE dermatan sulfate epimerase 2.70
EFHD1 EF‐hand domain family, member D1 18.39
EIF4ENIF1 eukaryotic translation initiation factor 4E nuclear import factor 1 2.10
ELOVL7 ELOVL family member 7, elongation of long chain fatty acids (yeast) 2.24
ELTD1 EGF, latrophilin and seven transmembrane domain containing 1 17.58
ENKUR enkurin, TRPC channel interacting protein 34.55
EpCAM epithelial cell adhesion molecule 24.20
EPN2 epsin 2 2.88
ESF1 similar to ABT1‐associated protein; ESF1, nucleolar pre‐rRNA
processing protein, homolog (S. cerevisiae) 11.30
EYA2 eyes absent homolog 2 (Drosophila) 77.76
FAM117A family with sequence similarity 117, member A 3.84
FAM196A chromosome 10 open reading frame 141 45.70
FAM92B family with sequence similarity 92, member B 106.17
FBLN2 fibulin 2 19.19
FER1L5 fer‐1‐like 5 (C. elegans) 25.03
FER1L6 fer‐1‐like 6 (C. elegans) 47.07
FGD2 FYVE, RhoGEF and PH domain containing 2 22.35
FGD5 FYVE, RhoGEF and PH domain containing 5 108.00
FGGY FGGY carbohydrate kinase domain containing 32.46
FMN1 formin 1 15.46
FRK fyn‐related kinase 2.10
FRMPD4 FERM and PDZ domain containing 4 6.22
FSTL4 follistatin‐like 4 8.83
FUZ fuzzy homolog (Drosophila) 2.41
GAB2 GRB2‐associated binding protein 2 2.75
GALNT12 UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide
N‐acetylgalactosaminyltransferase 12 (GalNAc‐T12) 4.15

GALNT8 UDP‐N‐acetyl‐alpha‐D‐galactosamine:polypeptide
N‐acetylgalactosaminyltransferase 8 (GalNAc‐T8) 94.33
GEM GTP binding protein overexpressed in skeletal muscle 112.38
GLIS1 GLIS family zinc finger 1 21.91
GPA33 glycoprotein A33 (transmembrane) 22.00
GPNMB glycoprotein (transmembrane) nmb 2.25
GPR155 G protein‐coupled receptor 155 33.09
GPR158 G protein‐coupled receptor 158 11.81
GPR176 G protein‐coupled receptor 176 12.62
GPR85 G protein‐coupled receptor 85 29.09
GRAMD3 GRAM domain containing 3 2.13
GRIK2 glutamate receptor, ionotropic, kainate 2 4.20
GULP1 GULP, engulfment adaptor PTB domain containing 1 2.90
HIF1A hypoxia inducible factor 1, alpha subunit (basic helix‐loop‐helix
transcription factor) 16.95
HOXD3 homeobox D3 2.29
HS3ST3B1 heparan sulfate (glucosamine) 3‐O‐sulfotransferase 3B1 2.05
IGFBP2 insulin‐like growth factor binding protein 2, 36kDa 197.93
IGFBP5 insulin‐like growth factor binding protein 5 14.42
IL17D interleukin 17D 3.58
IL1RAP interleukin 1 receptor accessory protein 33.89
INHBC inhibin, beta C 2.02
INMT indolethylamine N‐methyltransferase 13.90
ISPD notch1‐induced protein 3.24
ITGAL integrin, alpha L (antigen CD11A (p180), lymphocyte
function‐associated antigen 1; alpha polypeptide) 13.93
ITGBL1 integrin, beta‐like 1 (with EGF‐like repeat domains) 21.97
JAZF1 JAZF zinc finger 1 32.89
KAT6B K(lysine) acetyltransferase 6B 2.23
KATNAL1 katanin p60 subunit A‐like 1 2.63
KCNB1 potassium voltage‐gated channel, Shab‐related subfamily, member 1 131.34
KCNC1 potassium voltage‐gated channel, Shaw‐related subfamily, member 1 43.60
KCNK2 potassium channel, subfamily K, member 2 19.87
KCNQ5 potassium voltage‐gated channel, KQT‐like subfamily, member 5 11.18
KCTD12 potassium channel tetramerisation domain containing 12 2.29
KIAA1324 KIAA1324 7.80
KIAA1462 KIAA1462 9.85
KIAA1683 KIAA1683 2.34
KIF26B kinesin family member 26B 19.26
KIRREL3 kin of IRRE like 3 (Drosophila) 3.24
KLHL32 kelch‐like 32 (Drosophila) 6.06
KLHL7 kelch‐like 7 (Drosophila) 2.14
LEPR leptin receptor 15.92
LHCGR luteinizing hormone/choriogonadotropin receptor 14.78

LIMS2 LIM and senescent cell antigen‐like domains 2 47.15
LIPC lipase, hepatic 28.94
LIPN lipase, family member N 2.86
LOC10028703
6 hypothetical protein LOC100287036 6.17
LOC554223 hypothetical LOC554223 86.05
LOXL4 lysyl oxidase‐like 4 7.86
LRRC17 leucine rich repeat containing 17 14.84
LRRC4 leucine rich repeat containing 4 26.95
LY6D lymphocyte antigen 6 complex, locus D 28.96
MAF v‐maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) 47.09
MAGI2 membrane associated guanylate kinase, WW and PDZ domain
containing 2 4.51
MAP7D2 MAP7 domain containing 2 10.41
MB myoglobin 3.06
MED12L mediator complex subunit 12‐like 4.51
MLIP muscular LMNA‐interacting protein 30.43
MRAP2 melanocortin 2 receptor accessory protein 2 7.37
MSI1 musashi homolog 1 (Drosophila) 67.82
MXD1 MAX dimerization protein 1 11.17
NEGR1 neuronal growth regulator 1 3.43
NOS3 nitric oxide synthase 3 (endothelial cell) 4.36
NOSTRIN nitric oxide synthase trafficker 25.09
NOVA2 neuro‐oncological ventral antigen 2 185.06
NPL N‐acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) 11.92
NR5A2 nuclear receptor subfamily 5, group A, member 2 30.12
NYNRIN KIAA1305 38.45
P2RX3 purinergic receptor P2X, ligand‐gated ion channel, 3 60.93
P4HA1 prolyl 4‐hydroxylase, alpha polypeptide I 8.17
PAPPA2 pappalysin 2 46.30
PARK2 Parkinson disease (autosomal recessive, juvenile) 2, parkin 9.27
PBX3 pre‐B‐cell leukemia homeobox 3 88.94
PCDHA9 protocadherin alpha 9 2.16
PDGFC platelet derived growth factor C 5.64
PDK3 pyruvate dehydrogenase kinase, isozyme 3 2.65
PIWIL4 piwi‐like 4 (Drosophila) 20.51
PKD1L1 polycystic kidney disease 1 like 1 14.95
PLAG1 pleiomorphic adenoma gene 1 18.21
PLAT plasminogen activator, tissue 5.04
PLXDC1 plexin domain containing 1 2.58
PLXNA4 plexin A4 39.31
PPFIA1 protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF),
interacting protein (liprin), alpha 1 2.03
PPP1R1C protein phosphatase 1, regulatory (inhibitor) subunit 1C 223.65

PRELID2 PRELI domain containing 2 3.10
PRKAG2 protein kinase, AMP‐activated, gamma 2 non‐catalytic subunit 2.52
PRKAR2B protein kinase, cAMP‐dependent, regulatory, type II, beta 35.87
PRSS16 protease, serine, 16 (thymus) 2.11
PTAFR platelet‐activating factor receptor 11.49
PTPRC protein tyrosine phosphatase, receptor type, C 12.89
PTPRE protein tyrosine phosphatase, receptor type, E 2.37
PTPRR protein tyrosine phosphatase, receptor type, R 54.15
RAPGEF5 Rap guanine nucleotide exchange factor (GEF) 5 30.38
RARRES1 retinoic acid receptor responder (tazarotene induced) 1 33.08
RASSF4 Ras association (RalGDS/AF‐6) domain family member 4 4.32
RDX radixin 2.01
RFESD Rieske (Fe‐S) domain containing 2.48
RFX3 regulatory factor X, 3 (influences HLA class II expression) 23.46
RGAG1 retrotransposon gag domain containing 1 3.24
RGMA RGM domain family, member A 18.12
RIN2 Ras and Rab interactor 2 11.91
RNF182 ring finger protein 182 2.13
ROS1 c‐ros oncogene 1 , receptor tyrosine kinase 2.11
RPS6KB1 ribosomal protein S6 kinase, 70kDa, polypeptide 1 2.21
RTDR1 rhabdoid tumor deletion region gene 1 39.71
SALL2 sal‐like 2 (Drosophila) 6.24
SCG5 secretogranin V (7B2 protein) 36.45
SCN2B sodium channel, voltage‐gated, type II, beta 8.43
SDC2 syndecan 2 6.96
SEMA5A
sema domain, seven thrombospondin repeats (type 1 and type 1‐like),
transmembrane domain (TM) and short cytoplasmic domain,
(semaphorin) 5A
2.44
SERPINE3 serpin peptidase inhibitor, clade E (nexin, plasminogen activator
inhibitor type 1), member 3 21.66
SFMBT2 Scm‐like with four mbt domains 2 9.62
SGOL1 shugoshin‐like 1 (S. pombe) 9.40
SGOL2 shugoshin‐like 2 (S. pombe) 12.89
SIAH3 seven in absentia homolog 3 (Drosophila) 35.56
SLC14A2 solute carrier family 14 (urea transporter), member 2 9.59
SLC22A11 solute carrier family 22 (organic anion/urate transporter), member 11 111.92
SLC2A14 solute carrier family 2 (facilitated glucose transporter), member 14 35.10
SLC2A5 solute carrier family 2 (facilitated glucose/fructose transporter),
member 5 2.06
SLC8A2 solute carrier family 8 (sodium/calcium exchanger), member 2 32.53
SOX2 SRY (sex determining region Y)‐box 2 3.74
SPTY2D1 SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) 24.41
SWT1 Transcriptional protein SWT1 11.15
SYCP2 synaptonemal complex protein 2 2.00

SYCP2L synaptonemal complex protein 2‐like 6.67
SYN2 synapsin II 2.64
TACR1 tachykinin receptor 1 51.44
TBX19 T‐box 19 2.14
TEK TEK tyrosine kinase, endothelial 25.59
TFEC transcription factor EC 8.70
TNFRSF13B tumor necrosis factor receptor superfamily, member 13B 547.26
TOX thymocyte selection‐associated high mobility group box 168.89
TRAF5 TNF receptor‐associated factor 5 3.54
TREM1 triggering receptor expressed on myeloid cells 1 466.94
TRIM39 tripartite motif‐containing 39 16.63
TRIM71 tripartite motif‐containing 71 78.61
TSHZ2 teashirt zinc finger homeobox 2 7.55
TSPAN5 tetraspanin 5 3.51
UBE2W ubiquitin‐conjugating enzyme E2W (putative) 7.36
UCHL1 ubiquitin carboxyl‐terminal esterase L1 (ubiquitin thiolesterase) 44.44
WTH3DI RAB6C‐like 3.40
ZCCHC16 zinc finger, CCHC domain containing 16 14.47
ZDHHC22 zinc finger, DHHC‐type containing 22 41.79
ZFHX4 zinc finger homeobox 4 27.12
ZNF438 zinc finger protein 438 14.83
ZNF474 zinc finger protein 474 3.05
ZNF521 zinc finger protein 521 78.12
ZNF674 zinc finger family member 674 2.05
ZSCAN30 zinc finger and SCAN domain containing 30 2.19
Down‐regulated genes
ACSBG1 acyl‐CoA synthetase bubblegum family member 1 0.27
ADAP2 ArfGAP with dual PH domains 2 0.05
ADD3 adducin 3 (gamma) 0.26
ADRA1D adrenergic, alpha‐1D‐, receptor 0.01
AMACR C1q and tumor necrosis factor related protein 3; alpha‐methylacyl‐CoA
racemase 0.07
AMMECR1 Alport syndrome, mental retardation, midface hypoplasia and
elliptocytosis chromosomal region gene 1 0.11
ANGPTL5 angiopoietin‐like 5 0.34
ANKRD35 ankyrin repeat domain 35 0.03
APOL6 apolipoprotein L, 6 0.39
ARRB1 arrestin, beta 1 0.36
ASB14 ankyrin repeat and SOCS box‐containing 14 0.10
ATP8B4 ATPase, class I, type 8B, member 4 0.09
BCL2L11 BCL2‐like 11 (apoptosis facilitator) 0.31
BCMO1 beta‐carotene 15,15'‐monooxygenase 1 0.01
BST1 bone marrow stromal cell antigen 1 0.05
BTBD16 BTB (POZ) domain containing 16 0.05

BTC betacellulin 0.03
C12orf61 chromosome 12 open reading frame 61 0.38
C1orf194 chromosome 1 open reading frame 194 0.01
C22orf43 chromosome 22 open reading frame 43 0.41
C3orf43 chromosome 3 open reading frame 43 0.02
C5orf64 chromosome 5 open reading frame 64 0.31
C8orf46 chromosome 8 open reading frame 46 0.04
CAPZB capping protein (actin filament) muscle Z‐line, beta 0.47
CCL17 chemokine (C‐C motif) ligand 17 0.00
CD180 CD180 molecule 0.05
CDH22 cadherin‐like 22 0.08
CFI complement factor I 0.32
CFP complement factor properdin 0.03
CGREF1 cell growth regulator with EF‐hand domain 1 0.46
CHRNB2 cholinergic receptor, nicotinic, beta 2 (neuronal) 0.42
CTRC chymotrypsin C (caldecrin) 0.02
CXCL17 chemokine (C‐X‐C motif) ligand 17 0.01
DCLK1 doublecortin‐like kinase 1 0.36
DNAH3 dynein, axonemal, heavy chain 3 0.12
DPT dermatopontin 0.04
DPYSL5 dihydropyrimidinase‐like 5 0.05
DTX3L deltex 3‐like (Drosophila) 0.39
EFCAB5 EF‐hand calcium binding domain 5 0.04
EFTUD2 elongation factor Tu GTP binding domain containing 2 0.45
EIF2AK2 eukaryotic translation initiation factor 2‐alpha kinase 2 0.43
EIF4A1 eukaryotic translation initiation factor 4A, isoform 1 0.35
ELAC2 elaC homolog 2 (E. coli) 0.38
EPB41L3 erythrocyte membrane protein band 4.1‐like 3 0.05
EPHA4 EPH receptor A4 0.06
ERAP2 endoplasmic reticulum aminopeptidase 2 0.08
FAM178B family with sequence similarity 178, member B 0.05
FAM46C family with sequence similarity 46, member C 0.41
FAM53A family with sequence similarity 53, member A 0.09
FCER1G Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide 0.31
FIGF c‐fos induced growth factor (vascular endothelial growth factor D) 0.03
FRMD5 FERM domain containing 5 0.34
FRMD7 FERM domain containing 7 0.01
GABRR2 gamma‐aminobutyric acid (GABA) receptor, rho 2 0.03
GBP5 guanylate binding protein 5 0.07
GCNT7 glucosaminyl (N‐acetyl) transferase family member 7 0.37
GFAP glial fibrillary acidic protein 0.09
GFOD1 glucose‐fructose oxidoreductase domain containing 1 0.50
GIPC2 GIPC PDZ domain containing family, member 2 0.38
GLRB glycine receptor, beta 0.09

GMPR guanosine monophosphate reductase 0.14
GPR37 G protein‐coupled receptor 37 (endothelin receptor type B‐like) 0.04
GPR97 G protein‐coupled receptor 97 0.42
GRAP2 GRB2‐related adaptor protein 2 0.16
HIST1H2BL histone cluster 1, H2bl 0.17
HIST4H4
histone cluster 1, H4l; histone cluster 1, H4k; histone cluster 4, H4;
histone cluster 1, H4h; histone cluster 1, H4j; histone cluster 1, H4i;
histone cluster 1, H4d; histone cluster 1, H4c; histone cluster 1, H4f;
histone cluster 1, H4e; histone cluster 1, H4b; histone cluster 1, H4a;
histone cluster 2, H4a; histone cluster 2, H4b 0.38
HMP19 HMP19 protein 0.08
HOXD11 homeobox D11 0.04
HOXD13 homeobox D13 0.20
HPGD hydroxyprostaglandin dehydrogenase 15‐(NAD) 0.20
HRASLS2 HRAS‐like suppressor 2 0.02
IFIH1 interferon induced with helicase C domain 1 0.33
IFIT1 interferon‐induced protein with tetratricopeptide repeats 1 0.16
IFIT2 interferon‐induced protein with tetratricopeptide repeats 2 0.21
IFIT5 interferon‐induced protein with tetratricopeptide repeats 5 0.47
IL34 interleukin 34 0.50
ITIH5 inter‐alpha (globulin) inhibitor H5 0.08
ITK IL2‐inducible T‐cell kinase 0.02
KCNK12 potassium channel, subfamily K, member 12 0.04
KCNRG potassium channel regulator 0.05
KIAA0232 KIAA0232 0.44
KIAA1107 KIAA1107 0.41
KLRC4 killer cell lectin‐like receptor subfamily C, member 4 0.03
KLRC4‐KLRK1 KLRC4‐KLRK1 Read‐Through Transcript 0.04
LAMP3 lysosomal‐associated membrane protein 3 0.30
LCLAT1 lysocardiolipin acyltransferase 1 0.45
LDB3 LIM domain binding 3 0.10
LPHN3 latrophilin 3 0.03
LRP1B low density lipoprotein‐related protein 1B (deleted in tumors) 0.09
MAK male germ cell‐associated kinase 0.12
MARK2 MAP/microtubule affinity‐regulating kinase 2 0.49
MMRN1 multimerin 1 0.03
MYBPHL myosin binding protein H‐like 0.01
MYO9B myosin IXB 0.34
NIPAL2 NIPA‐like domain containing 2 0.20
NME5 non‐metastatic cells 5, protein expressed in (nucleoside‐diphosphate
kinase) 0.01
NR0B2 nuclear receptor subfamily 0, group B, member 2 0.01
NR1D2 nuclear receptor subfamily 1, group D, member 2 0.08
NR6A1 nuclear receptor subfamily 6, group A, member 1 0.39

NT5C1B‐RDH1
4 NT5C1B‐RDH14 Read‐Through Transcript
0.09
OTX1 orthodenticle homeobox 1 0.43
PABPC4L poly(A) binding protein, cytoplasmic 4‐like 0.00
PADI4 peptidyl arginine deiminase, type IV 0.02
PARD3B par‐3 partitioning defective 3 homolog B (C. elegans) 0.09
PARP10 poly (ADP‐ribose) polymerase family, member 10 0.49
PARP12 poly (ADP‐ribose) polymerase family, member 12 0.41
PARP14 poly (ADP‐ribose) polymerase family, member 14 0.43
PCSK5 proprotein convertase subtilisin/kexin type 5 0.06
PCSK6 proprotein convertase subtilisin/kexin type 6 0.50
PGPEP1L pyroglutamyl‐peptidase 1‐like 0.04
PIK3AP1 phosphoinositide‐3‐kinase adaptor protein 1 0.39
PLVAP plasmalemma vesicle associated protein 0.03
PMF1‐BGLAP PMF1‐BGLAP Read‐Through Transcript 0.46
POC1B‐GALN
T4 POC1B‐GALNT4 Read‐Through Transcript
0.12
POF1B premature ovarian failure, 1B 0.02
PPARD peroxisome proliferator‐activated receptor delta 0.41
PPP6C protein phosphatase 6, catalytic subunit 0.50
PRICKLE1 prickle homolog 1 (Drosophila) 0.06
PRPH2 peripherin 2 (retinal degeneration, slow) 0.02
PRR5‐ARHGA
P8
Rho GTPase activating protein 8; proline rich 5 (renal); PRR5‐ARHGAP8
fusion 0.05
PSMD1 proteasome (prosome, macropain) 26S subunit, non‐ATPase, 1 0.06
PTPRB protein tyrosine phosphatase, receptor type, B 0.13
RAB9B RAB9B, member RAS oncogene family 0.02
RAG2 recombination activating gene 2 0.02
RAP1GDS1 RAP1, GTP‐GDP dissociation stimulator 1 0.50
RARRES3 retinoic acid receptor responder (tazarotene induced) 3 0.30
RBP1 retinol binding protein 1, cellular 0.36
RNF183 ring finger protein 183 0.01
SAMD5 sterile alpha motif domain containing 5 0.13
SAMHD1 SAM domain and HD domain 1 0.33
SDHD
similar to succinate dehydrogenase complex, subunit D, integral
membrane protein; succinate dehydrogenase complex, subunit D,
integral membrane protein 0.06
SECTM1 secreted and transmembrane 1 0.43
SEMA6A sema domain, transmembrane domain (TM), and cytoplasmic domain,
(semaphorin) 6A 0.49
SGPP2 sphingosine‐1‐phosphate phosphotase 2 0.03
SIDT1 SID1 transmembrane family, member 1 0.01
SLC25A13 solute carrier family 25, member 13 (citrin) 0.30
SLC7A7 solute carrier family 7 (cationic amino acid transporter, y+ system), 0.09

member 7
SLC7A8 solute carrier family 7 (cationic amino acid transporter, y+ system),
member 8 0.08
SLFN11 schlafen family member 11 0.25
SRL sarcalumenin 0.48
ST18 suppression of tumorigenicity 18 (breast carcinoma) (zinc finger
protein) 0.01
ST3GAL5 ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 5 0.44
ST3GAL6 ST3 beta‐galactoside alpha‐2,3‐sialyltransferase 6 0.06
STOX1 storkhead box 1 0.48
STX19 syntaxin 19 0.06
STXBP5L syntaxin binding protein 5‐like 0.17
SYNJ2BP‐COX
16 SYNJ2BP‐COX16 Read‐Through Transcript
0.04
SYNPO2 synaptopodin 2 0.45
TAS2R30 taste receptor, type 2, member 30 0.03
TBRG4 transforming growth factor beta regulator 4 0.46
TEX19 testis expressed 19 0.48
TLR3 toll‐like receptor 3 0.40
TMC6 transmembrane channel‐like 6 0.47
TMEM232 hypothetical protein LOC642987 0.47
TMEM51 transmembrane protein 51 0.04
TMPRSS6 transmembrane protease, serine 6 0.31
TNFRSF8 tumor necrosis factor receptor superfamily, member 8 0.05
TNFSF10 tumor necrosis factor (ligand) superfamily, member 10 0.47
TOMM40 translocase of outer mitochondrial membrane 40 homolog (yeast) 0.41
TRANK1 lupus brain antigen 1 0.24
TRIM14 tripartite motif‐containing 14 0.41
TRIM2 tripartite motif‐containing 2 0.47
TRIM21 tripartite motif‐containing 21 0.41
TTC23L tetratricopeptide repeat domain 23‐like 0.01
ULK2 unc‐51‐like kinase 2 (C. elegans) 0.29
WDR86 WD repeat domain 86 0.38
WDR93 WD repeat domain 93 0.08
WNT2B wingless‐type MMTV integration site family, member 2B 0.03
XYLT1 xylosyltransferase I 0.11
YTHDF2 YTH domain family, member 2 0.47
ZFAT zinc finger and AT hook domain containing 0.44
ZKSCAN5 zinc finger with KRAB and SCAN domains 5 0.45
ZNF512 zinc finger protein 512 0.49

Supplementary Table 9. Primers sequences
Name Sequence
Primers for constructing down-regulated miRNAs in Hep-12 cells
miR-122: PF: 5'CGGGATCCACAGGAAGTTTTTGCAGAAG
PR: 5'CCGCTCGAGAGCATGTGAGAGGCAGGGTTC
miR-126: PF: 5'CGGGATCCTATGCCGCCTAAGTACGTC
PR: 5'CCGCTCGAGAGCCTCATATCAGCCAAGAAG
miR-130a PF: 5'CGGGATCCAGGGGGTTGGTGAAGAAGG
PR: 5'CCGCTCGAGTTTCACAGTAACGGAGGCAG
miR-149 PF: 5'CGGGATCCTCCTGCCTTTGACTGCCG
PR: 5'CCGCTCGAGTCAGCCACCTCTCACACC
miR-182 PF: 5'CGGAATTCTGCAGGTTACAGATATGAGG
PF: 5'CGGGATCCTGCAGGTTACAGATATGAG
miR-183 PF: 5'CGGGATCCTGGGGCATGTGGATCTTGTG
PR: 5'CCGCTCGAGTCCTGCTTTTCCCATATTG
miR-200b PF: 5'CGGGATCCTCAGACAAGGCAGCACGTG
PR: 5'CCGCTCGAGTTGCATTCCGGGGTCTCTG
miR-221 PF: 5'CGGGATCCAGTTTATCTATCCGACCTTC
PR: 5'CCGCTCGAGACTAGGCTGGTGTGTGAGAC
miR-222 PF: 5'CGGGATCCAGGAATCATGTATGCTGTAG
PR: 5'CCGCTCGAGACTTAACACCCTAGAACTTG
miR-224 PF: 5'CGGGATCCTACTACCTTTGACTTGGGAG
PR: 5'CCGCTCGAGAAGAGTTTGAGACCAGTCC
miR-23a PF: 5'CGGGATCCAAGCATCTGGGGACCTGGAG
PR: 5'CCGCTCGAGACTTAGCCACTGTGAACACG
miR-23b PF: 5'CGGGATCCTCATCTCTGGAGTCCCTTTG
PR: 5'CCGCTCGAGTTGTTAGAGAGGTCATCGCTG
miR-28 PF: 5'CGGGATCCAGAAGTGTCCTGAATGTTTAC
PR: 5'CCGCTCGAGTGACAAAGAGGAGCTAGAAG

miR-29a PF: 5'CGGGATCCTTGTACAGGATATCGCATTG
PR: 5'CCGCTCGAGTCAACTTAGAAACTGGAAGA
miR-31 PF: 5'CGGGATCCACCTCCTGTGCCTAACTAC
PR: 5'CCGCTCGAGAGGTGTGTCCAAGGAATAG
miR-424 PF: 5'CGGGATCCTCGCTCTGCTTTTCACCCAC
PR: 5'CCGCTCGAGAATACTGCCCTCCCCGGAC
miR-452 PF: 5'CGGGATCCTCACATTCCTTTCCAGTTTC
PR: 5'CCGCTCGAGACTGGATTGGTTCTGGGTATG
miR-455
PF: 5'CGGGATCCAGCCGCTGTTAGTTAATGCC
PR: 5'CCGCTCGAGAATGTAGCCCTAGGAAAGAG
miR-663 PF: 5'CGGAATTCTGGACGAGAATCACGAGCG
PR: 5'CCGCTCGAGACCAACCGACGTAAAGCC
miR-99b PF: 5'CGGGATCCAGGCTTTGGATCTGGACTC
PR: 5'CCGCTCGAGACAGACCCGACAGACAGAC
Let-7a PF: 5'CGGGATCCAAGAAGCAAAAGGTTTCCCC
PR: 5'CCGCTCGAGAGCGGATTCAGATAACCAAG
miR-21 PF: 5'CGGGATCCGATTGAACTTGTTCATTTTG
PR: 5'CCGCTCGAGAAGACTATCCCCATTTCTCCA
miR-34a PF: 5'CGGATCCCGGGCTGGAGAGAC
PR: 5'CCGCTCGAGCCTGTGCCTTTTTCCTTCC
Let-7b PF: 5’CGGGATCCATTGAATCTAGCTCCCAGATGC
PR: 5’CCGCTCGAGCAGGAAGTGAGAGGAACAGC
Let-7d PF: 5’CGGGATCCAGGAGGAACAGCTGAGAGTC
PR: 5’CCGCTCGAGTCGTTTGTGACTTGAGGTTG
Let-7e PF: 5’CGGGATCCAGGAGGGGATGCAGGGACAAG
PR: 5’CCGCTCGAGACCCGTAGAACCGACCTTGC
Let-7f PF: 5’CGGGATCCAGTGCAGCATTTGTTTATGG
PR: 5’ CCGCTCGAGCTATGAAGATTTCCTGAAGG
Let-7g PF: 5’CGGGATCCACTGGTTTATTTCTTCTGTTG

PR: 5’CCGCTCGAGAGTCATCCCAGGGGTTCACAG
Let-7c PF: 5’CACGGATCGACATTTTACGTGACCTATC
PR: 5’CACCTCGAGCATGGAGTGACAACCCATTAG
miR-125a PF: 5’CGGGATCCAGTAGGTTGTGAAGCAGAGG
PR: 5’CCGCTCGAGTCTGACTCCCTCTTATTCTG
Primers for constructing up-regulated miRNAs in Hep-12 cells
miR-10b PF:5'CGGGATCC AGTACCCCAAAACCTAGCC
PR:5'CCGCTCGAG TGTCGCCAAGCCCATTAGG
miR-1224
PF:5'CGGGATCC TGATGATCCCCACTGCAGG
PR:5'CCGCTCGAG ACTGTCACCCTGGCCAAC
miR-124-1
PF:5'CGGGATCC ATGGGGGAGAACAAAGAGC
PR:5'CCGCTCGAG TGCATCTCTAAGCCCCTGTC
miR-124-2
PF:5'CGGGATCC AACAGACTAATTAACAGCGG
PR:5'CCGCTCGAG TTATGCGGCAAGAGATGGG
miR-124-3
PF:5'CGGGATCC AAAGGGGAGAAGTGTGGGC
PR:5'CCGCTCGAG CATTGTTCGCCGGATTTGTC
miR-125b-1
PF:5'CGGGATCC ACCATACCACCTGTTTGTTG
PR:5'CCGCTCGAG TATACTCAATCACCTCAGAC
miR-125b-2
PF:5'CGGGATCC ATGGTCGTCGTGATTACTC
PR:5'CCGCTCGAG AGCATTGTTCTTTTCTCCTAG
miR-130b
PF:5'CGGGATCC ATGATTCTCCAGCTCTGCAC
PR:5'CCGCTCGAG AATTCGCTCCCTTCTCCATG
miR-132
PF:5'CGGGATCC ACAGTCTCCAGTCACGGCC
PR:5'CCGCTCGAG ACGTGGGATCTTGACTCGC
miR-137
PF:5'CGGGATCC TTTATGGTCCCGGTCAAGC
PR:5'CCGCTCGAG AACCCATTTTCCAGTCCTG
miR-148a
PF:5'CGGGATCC TTCCATTATCGGTCGCATCC
PR:5'CCGCTCGAG ATTGGTCAAGTTCCTCGTGC
miR-148b PF:5'CGGGATCC AGGACAATGGACTAGGCTT

PR:5'CCGCTCGAG TCTGTCTAAGTCACCCAATC
miR-192 PF:5'CGGGATCC AGTGGGGCTGCTGTTATCT
PR:5'CCGCTCGAG TCTGCTGACTGCTGGACAC
miR-194-2
PF:5'CGGGATCC ATGATAAGAAGCCTCGGTG
PR:5'CCGCTCGAG AATTCATAGGTCAGAGCCC
miR-204
PF:5'CGGGATCC TGATGGAAAGGAGGGTGGG
PR:5'CCGCTCGAG TTATGGGACAGTTATGGGCC
miR-216a
PF:5'CGGGATCC TGTGAATTCAGCAAGAGGAG
PR:5'CCGCTCGAG TCCTTTCCAGCACAACAGC
miR-217
PF:5'CGGGATCC ATCTCTCTTGGTAAATTGGG
PR:5'CCGCTCGAG AGAACGGTGGTAAATGATAG
miR-375 PF:5'CGGGATCC AGTATAAGAGCACACGGAGC
PR:5'CCGCTCGAG TTACGACGCAGAATGGAGC
miR-483 PF:5'CGGGATCC ATTTGGGGGTAGGAAGTGG
PR:5'CCGCTCGAG AAATGTGTGGGATGTGAGC
miR-512-1
PF:5'CGGGATCC TGTTATTCAGGCTGGGGAC
PR:5'CCGCTCGAG TTAGCTTTTACCTGCAGTGG
miR-512-2
PF:5'CGGGATCC AGCAACATTCAGAAGTCAG
PR:5'CCGCTCGAG TTTATGGCGCAGAACAAGC
miR-515
PF:5'CGGGATCC AGTAGAGACGGAGTTTCAC
PR:5'CCGCTCGAG ACAGAACCCCAACATCATG
miR-516a
PF:5'CGGGATCC CTAACCTGCTGATTCTTTGA
PR:5'CCGCTCGAG TGTATTACCAAGATCAGCGT
miR-516b-1
PF:5'CGGGATCC TCCAGGAAACATGCAAACAG
PR:5'CCGCTCGAG AGATCTAGGTTGGAAGTGC
miR-517a PF:5'CGGGATCC TGAACCCAGGAGGCAGAAG
PR:5'CCGCTCGAG CTGATGATAAGCACGCTCTG
miR-517b
PF:5'CGGGATCCAGGCTAAGGCTTGAGAATG
PR:5'CCGCTCGAGAGCCCAAGAGATCTCGGTT

miR-517c
PF:5'CGGGATCCAGAAGGAGATTGCAGTGAG
PR:5'CCGCTCGAGAACCCCAACATCATGCACG
miR-518b
PF:5'CGGGATCCACCTGCTGAGCCTTTGAAG
PR:5'CCGCTCGAGAGCCCAAGAGATCTAGGTAG
miR-518d:
PF:5'CGGGATCCGAAGCAAAGAACCAGAGATG
PR:5'CCGCTCGAGAAGAGACCTAGGTTGGAAG
miR-518e PF:5'CGGGATCCAGTGGTTTATGTTCTGGATTC
PR:5'CCGCTCGAGTGAACCCAAGAGATCTAGG
miR-518f PF:5'CGGGATCCACCTGCTGATCCTTTGAAG
PR:5'CCGCTCGAGAACACCAACATCATCCACG
miR-519a-1 PF:5'CGGGATCCTTGAGGGAAGGAACTGGAG
PR:5'CCGCTCGAGTCAGAAGCCCAACATCATC
miR-519a-2 PF:5'CGGGATCCAGAAATCCAGCCTAATCCTC
PR:5'CCGCTCGAGTGAGCCCAAGAGATCTAGG
miR-519c PF 5'CGGGATCCAACAAGGAACTGGAGATGG
PR:5'CCGCTCGAGAAGAGAGCTAGGTTGGAAG
miR-519d PF:5'CGGGATCCACCTGCAGAGCCTTTGAAG
PR:5'CCGCTCGAGACTCCCAACATCATCCAAG
miR-519e PF 5'CGGGATCCAAGCAAGGAACTGGAGATG
PR:5'CCGCTCGAGATGCGTTACCAAGATCAGC
miR-520a PF:5'CGGGATCCAAGAAGGAACTGGAGATGG
PR:5'CCGCTCGAGAGAACCCCAACATCATCCG
miR-520c PF:5'CGGGATCCGAAGGAACTGGAGATGGTC
PR:5'CCGCTCGAGAGAACCCCAACATCATCCG
miR-520g
PF:5'CGGGATCCTGTAAGGTATGAGTAGTAGG
PR:5'CCGCTCGAGAACCCCAACATCATCCAAG
miR-520h PF:5'CGGGATCCACCTGCTGAGCCTTTGAAG
PR:5'CCGCTCGAGAACCCCAACATCATCCAAG
miR-522 PF:5'CGGGATCCAAGCAAGGAACTGGAGATG

PR:5'CCGCTCGAGAAGAGGTCTAGGTTGGAAG
miR-525 PF:5'CGGGATCCAAGGAACTGGAGATGGTCG
PR:5'CCGCTCGAGACCCACCATCATCCAAGTC
miR-629 PF:5'CGGGATCCAATCCCTGCTTCCACTTTG
PR:5'CCGCTCGAGAACAATGCCACACAGATGC
miR-7-1 PF:5'CGGGATCCAACTTCTCATTTCTCTGGTG
PR:5'CCGCTCGAGATCGGACATTAGTAGAACAG
miR-7-2 PF:5'CGGGATCCTCTGCCGATGGGTGTTTAG
PR:5'CCGCTCGAGATGAACTTAGTAATAGGAGCG
miR-7-3 PF:5'CGGGATCCACGAGTGATGTGACAAACC
PR:5'CCGCTCGAGAACCGGGAATGGAAAACAG
miR-708 PF:5'CGGGATCCACTGGTGACTTCATTCCCC
PR:5'CCGCTCGAGTTGAGTTGGCAGTGCTGTG
miR-886 PF:5'CGGGATCCAGGACACGTTAGCAGGAC
PR:5'CCGCTCGAGATACTTTCTGTCACACCTTC
miR-9-2 PF:5'CGGGATCCACACACAAACAGAAACCAGG
PR:5'CCGCTCGAGTGCAACAACCCCTCTCAAG
miR-95 PF:5'CGGGATCCACCAGTTCCATTTGCACAC
PR:5'CCGCTCGAGACGCTTCTCCAGGTCAGG
miR-99a PF:5'CGGGATCCTGCATCCTTAGAACTCAGC
PR:5'CCGCTCGAGTCGAACTTGAATGGGTGTG
PolyA tailing qRT-PCR primers
OligodT adapter 5'GCGAGCACAGAATTAATACGACTCACTATAGGTTTTT
TTTTTTTVN
Reverse primer 5' GCGAGCACAGAATTAATACGAC
Let-7c forward 5' GTTGAGGTAGTAGGTTGTATGGTT
miR-200b forward 5’TAATACTGCCTGGTAATGATGAC
miR-222 forward 5'AGCTACATCTGGCTACTGGGT
miR-424 forward 5'GCAGCAGCAATTCATGTTTTGAA

Primers for construction of PBX3 3’UTR and corresponding mutation
PBX3- 3’UTR-PF 5' TGCTCTAGAccggTCTCTGGCCACACTTTTCCCTG
PBX3- 3’UTR-PR 5’CGGAATTCATGCATACATATAACTACCACGAGGAAG
AC
PBX3-200b-mut-PR 5’GACGTCAAAATCTGGACACATCACAATTAGTCGCTG
CTT
PBX3-200b-mut-PF
5’TGTCCAGATTTTGACGTCCAGTACTGTATATTTCACCC
TG
PBX3-222-mut-PF 5’AAGACGAAATTTCAATGTATTCCGCGATCTAGAGTGC
TCACTTACTACCTC
PBX3-222-mut- PR: 5’GATCGCGGAATACATTGAAATTTCGTCTTTTTGTATG
AGTTCGCATGAGG
PBX3-424-mut2-PF 5’AGACGAAGATACTGTAGTAGTATAGAGGTGAAAACG
AGATTG
PBX3-424-mut2-PR 5’ACTACTACAGTATCTTCGTCTCTCAGAACAATC