Suppl. Fig. 1
1
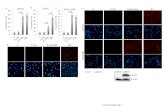
0 10 20 30 40 50 60 0 5 10 15 D ex [nM ] % Sub-G1 PD1.6 Cells 0µM GSNO 50µM GSNO 75µM GSNO 100µM GSNO Suppl. Fig. 1 y = 1.01x + 3.34 y = 2.33x + 7.12 y = 3.20x + 13.57 y = 5.03x + 1 7.51 Figure Legend. NO· synergizes with GC in inducing apoptosis of PD1.6 cells. PD1.6 were incubated o.n. with various concentrations of Dexamethasone (Dex) and/or S-Nitrosoglutathione (GSNO). The extent of apoptosis was measured by cell cycle analysis. A representative of eight separate experiments is shown. Statistical bars shown in the graphs correspond to mean +/- SD. The equation of the linear curve between 1-5nM Dex was calculated. The differences in the slopes indicate a synergistic effect.
-
Upload
satinka-herrera -
Category
Documents
-
view
20 -
download
2
description
Suppl. Fig. 1. y = 5.03x + 17.51. y = 3.20x + 13.57. y = 2.33x + 7.12. y = 1.01x + 3.34. - PowerPoint PPT Presentation
Transcript of Suppl. Fig. 1

0
10
20
30
40
50
60
0 5 10 15Dex [nM]
% S
ub-G
1 P
D1.
6 C
ells
0µM GSNO
50µM GSNO
75µM GSNO
100µM GSNO
Suppl. Fig. 1
y = 1.01x + 3.34
y = 2.33x + 7.12
y = 3
.20x
+ 1
3.57y
= 5.
03x
+ 17
.51
Figure Legend. NO· synergizes with GC in inducing apoptosis of PD1.6 cells. PD1.6 were incubated o.n. with various concentrations of Dexamethasone (Dex) and/or S-Nitrosoglutathione (GSNO). The extent of apoptosis was measured by cell cycle analysis. A representative of eight separate experiments is shown. Statistical bars shown in the graphs correspond to mean +/- SD. The equation of the linear curve between 1-5nM Dex was calculated. The differences in the slopes indicate a synergistic effect.