Poster Print Size: Nanopore Sensors And Signal Processing … › ... › 08 ›...
Transcript of Poster Print Size: Nanopore Sensors And Signal Processing … › ... › 08 ›...

Poster Print Size: This poster template is 24” high by 36” wide. It can be used to print any poster with a 2:3 aspect ratio including 36x54 and 48x72.
Placeholders: The various elements included in this poster are ones we often see in medical, research, and scientific posters. Feel free to edit, move, add, and delete items, or change the layout to suit your needs. Always check with your conference organizer for specific requirements.
Image Quality: You can place digital photos or logo art in your poster file by selecting the Insert, Picture command, or by using standard copy & paste. For best results, all graphic elements should be at least 150-200 pixels per inch in their final printed size. For instance, a 1600 x 1200 pixel photo will usually look fine up to 8“-10” wide on your printed poster.
To preview the print quality of images, select a magnification of 100% when previewing your poster. This will give you a good idea of what it will look like in print. If you are laying out a large poster and using half-scale dimensions, be sure to preview your graphics at 200% to see them at their final printed size.
Please note that graphics from websites (such as the logo on your hospital's or university's home page) will only be 72dpi and not suitable for printing.
[This sidebar area does not print.]
Change Color Theme: This template is designed to use the built-in color themes in the newer versions of PowerPoint.
To change the color theme, select the Design tab, then select the Colors drop-down list.
The default color theme for this template is “Office”, so you can always return to that after trying some of the alternatives.
Printing Your Poster: Once your poster file is ready, visit www.genigraphics.com to order a high-quality, affordable poster print. Every order receives a free design review and we can deliver as fast as next business day within the US and Canada.
Genigraphics® has been producing output from PowerPoint® longer than anyone in the industry; dating back to when we helped Microsoft® design the PowerPoint® software.
US and Canada: 1-800-790-4001
Email: [email protected]
[This sidebar area does not print.]
Nanopore Sensors And Signal Processing Melvin L Bowers III ,REU Student, Shepherd University
Graduate Mentor:Uday Shankar Shanthamallu Faculty Advisors: Dr. Michael Goryll, Dr. Andreas Spanias
MOTIVATION
Ion channels and nanopores can be modified and used to sequence DNA or detect specific disease relevant or toxic molecules [1-3].
PROBLEM STATEMENT
Signals are small and signatures are difficult to distinguish from artifacts.
System-related noise often masks transitions between different states of the ion channel or nanopore sensor.
Noise filtering is performed in the analog domain, often distorting the original shape of the signal.
Analog filtering can impact event classification, leading to false positives or negatives.
METHOD
Simulate ion channel signals using QuB to determine the expected signal behavior and analyze specific features in the signal.
Use wavelet decomposition [4].
Reduce white noise by picking only the most prominent wavelet components for re-synthesis.
Use the Signal-to-noise ratio (SNR) values to compare the efficiency of noise reduction.
:
FINAL RESULTS
Imported files into MATLAB and utilized MATLAB’S library of wavelets to perform an analysis of the simulated signal.
Utilizing different wavelets we determined the wavelet and coefficients that would result in the highest fidelity of the re-synthesized signal compared to the original.
The optimum wavelet and order was quantified by computing the SNR value for every de-noised signal which was then subtracted from the original SNR. The SNR of the de-noised signal includes the complete signal, taking into account possible over- and undershoot events at the state transitions, which are considered detrimental to subsequent classification.
Sensor Signal and Information Processing Center http://sensip.asu.edu
SenSIP Algorithms and Devices REU
ABSTRACT
REFERENCES
Ion channels are involved with everything from pain to disease in any organism.
Signal processing can be applied to de-noise ion-channel generated signals.
We demonstrate the use of Wavelet analysis and re-synthesis for de-noising, selecting the optimal wavelet base for the particular nature of ion-channel and nanopore signals.
[1] J. Clarke, H. Bayley et al., Nature Nanotechnology 4, 265–270 (2009).
[2] I. M. Derrington, E. Manrao, J. H. Gundlach et al., PNAS 107 (37), 16060-16065 (2010).
[3] S. Choi, M. Goryll, L.Y.M. Sin,et al. Microfluid Nanofluid (2011) 10: 231. doi:10.1007/s10404-010-0638-8
[4] B. Konnanath, P. Sattigeri, T. Mathew, A. Spanias, S.Prasad, M. Goryll, T. J. Thornton , P. Knee, ICANN09
This material is based upon work supported by the National Science Foundation under Grant No. CNS 1659871 REU Site: Sensors, Signal and Information Processing Devices and Algorithms.
Number of wavelet
orders used
Wavelet type
SNR of original signal (dB)
SNR of denoised
signal (dB)
SNR gained or lost from
baseline (dB)
5 Haar 18.29 19.22 0.93 6 Sym 18.29 18.00 -0.29 5 Coif 18.29 21.02 2.73 5 Bior 18.29 19.22 0.93 5 Rbio 18.29 19.22 0.93 4 Dmey 18.29 23.72 5.43 4 Fk 18.29 21.88 3.59
Original Signal
1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 0
Time (ms)
-0.4
1.4
1.2
1
0.8
0.4
0
0.6
0.2
-0.2
Am
plit
ud
e (
a.u
.)
Denoised DMEY
1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 0
Time (ms)
1
0.4
0
0.6
0.2
0.8
Am
plit
ud
e (
a.u
.)
Denoised HAAR
1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 0
Time (ms)
0
0.8
0.7
1
0.6
0.4
0.5
0.3
0.1
Am
plit
ud
e (
a.u
.)
0.2
0.9
Am
plit
ud
e (
a.u
.)
Denoised SYM
1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 0
Time (ms)
-0.2
1.2
1
0.8
0.4
0
0.6
0.2
Am
plit
ud
e (
a.u
.)
Denoised FK
1000 2000 3000 4000 5000 6000 7000 8000 9000 10000 0
Time (ms)
1
0.4
0
0.6
0.2
0.8
Am
plit
ud
e (
a.u
.)
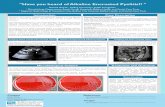
Representative signals from biological nanopore sensors used for DNA sequencing: (a) using an a-HL pore [1] and (b) using an MspA pore [2].
Simulated ion channel signal with added white noise using the QuB software package. The red trace shows the idealized signal using a Hidden Markov Model for state classification. (https://qub.mandelics.com/online)