More Scripting Techniques Scripting Process Example Script 1.
Pipeline Scripting for the Parallel Alignment of Genomic Short Sequence Reads
-
Upload
adam-bradley -
Category
Documents
-
view
7 -
download
1
Transcript of Pipeline Scripting for the Parallel Alignment of Genomic Short Sequence Reads

Pipeline Scripting for the Parallel Alignment of Genomic Short Sequence Reads
Adam Bradley, Kevin Drees, Paul Keim, Jeffrey FosterNorthern Arizona University Center for Microbial Genetics and Genomics
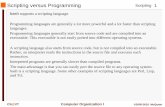
• Python pipeline script developed to input reference fasta and paired-end Illumina fastq.gz read files into alignment scripts
• locates the read files and reference sequence file in the working directory using regular expressions
• open source scripts integrated into pipeline
• bwa-0.7.5a: index reference fasta and produce raw alignment
• picard-tools-1.83: sort reads in alignment by reference coordinate, collect alignment statistics, remove duplicate reads, index alignment relative to reference
• samtools-0.1.19: remove reads mapping to more than one locus, baq analysis
• Commands submitted as jobs to pbs_server
Methods
• Single Nucleotide Polymorphisms (SNPs) in whole-genome sequences can be used to produce high-resolution phylogenetic trees, which illustrate the genetic relationships between organisms.
• Illumina NGS sequencing produces paired short sequence reads (100-250 bp) that must be assembled together to perform such genomic analyses.
• To find SNPs in whole-genome sequences, sequence reads can be aligned to a complete reference genome.
• Multiple software scripts are involved in the production of an alignment. It is cumbersome to run these scripts manually, particularly when aligning multiple sequences
• Our goal is to pipe (i.e., connect) the alignment programs together into a “pipeline,” which will reduce user effort and allow for parallel processing.
Introduction
The recent advent of Next-Generation Sequencing (NGS) technologies have allowed for the rapid and accurate sequencing of organisms' entire genomes, from small viral genomes to large animal genomes. Analysis of Single Nucleotide Polymorphisms (SNPs), or point mutations, across genomes allow geneticists to determine evolutionary relationships between organisms. However, NGS sequencers produce billions of short sequence "reads" that must be reassembled into their original contiguous sequence before being used in a SNP analysis. One method of doing so is called an alignment, in which reads are mapped to a completed reference sequence. Alignments often involve the use of several software scripts, and post-processing is usually needed to make the output compatible with downstream software. Furthermore, SNP analyses often include large numbers of samples to process. We developed a script in the Python programming language to connect the various scripts used in sequence alignments into a single process. The script is also able to process data from multiple samples in parallel, saving time and efficiently using available computer resources.
Abstract
• Pipeline currently indexes reference and produces raw alignments of multiple samples in parallel with bwa
• Downstream processes in progress
• Future versions will incorporate:
• average alignment depth of coverage and warning flags indicating poor alignment (Genome Analysis Toolkit DiagnoseTargets)
• % reference covered by aligned reads
• editing bam alignment headers to include more information about the sequencing run
Discussion
Funding was provided by the National Institutes of Health Bridges to Baccalaureate program and the National Cancer Institute Native American Cancer Prevention program.
Acknowledgements
Results
• Input reference file in fasta format
• Input paired-end Illumina reads files in fastq or gzipped fastq.gz format
• Output alignment must be in bam format, and have the following characteristics:
• reads sorted by reference coordinate
• perfectly duplicated reads removed
• reads mapping to more than one locus on reference removed
• Alignment index file in bai format must be output, for use by alignment viewers and SNP callers downstream
• Output must include mapping statistics and read insert size statistics
• Must align reads from multiple samples in parallel
• Subprocesses must be submitted as pbs jobs to a batch queuing server
Design SpecificationsFigure 2. Alignment of Brucella abortus 10-1086 to Brucella abortus 2308 reference, displayed with Tablet 1.13.05.17
picard SortSam
bwa index
reference.fasta
samtools view
bam alignment bai index
bwa mem
Illumina paired- end .fastq read files
picard Collect Insert Size Metrics
picard Collect Alignment Summary
Metrics
picard MarkDuplicates
samtools calmdpicard Build Bam Index
metrics files
Figure 1. Pipeline flow diagram