Part 11 lecture (eukaryotic nucleus)
-
Upload
eric-jones-uy -
Category
Documents
-
view
223 -
download
0
Transcript of Part 11 lecture (eukaryotic nucleus)
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
1/29
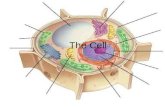
EUKARYOTIC NUCLEUS
Nucleusresponsible for the storage and utilization of genetic
informationStructure:
1. nuclear envelope
>the most important feature that distinguishes eukaryotesfrom prokaryotes>serve as a barrier that prevents ions, solutes, andmacromolecules from passing freely between the nucleusand cytoplasm>consists of 2 cellular membrane:a. outer membrane
- generally studded with ribosomes and continuous withthe membrane of the RER
b. inner membrane- bound by integral proteins to a thin filamentous
meshwork called nuclear lamina
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
2/29
> contains several thousand nuclear pores, which plays arole in nucleocytoplasmic exchange
2. Chromosomes> threadlike strands that are composed of the nuclear DNAof eukaryotic cells and are the carriers of geneticinformation
3. Nucleoli> irregularly shaped electron-dense structures thatfunction in the synthesis of ribosomal RNA
4. Nucleoplasm> the fluid substance in which the solutes of the nucleusare dissolved
5. Nuclear matrix> protein containing fibrillar network
Genomecollective term for the entire genetic information in every
nucleated eukaryotic cell
I. Physical and Structural Organization of the GenomeA. Deoxyribonucleic acid (DNA)
Chain of nucleotides
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
3/29
Nucleotides
Nucleoside Phosphate backbone
Nitrogenous base Sugar
purine pyrimidine 2-deoxy-D-ribose D-ribose(DNA) (RNA)
adenine guanine thymine cytosine(DNA)
uracil (for RNA)
Double right hand helix> proposed by Watson and Crick> makes one complete turn every 10 base pairs (34 A 0 or 34nm)> spaces between adjacent turns of the helix form 2 groovesof different width, a wider major groove and a more narrowminor groove
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
4/29
Double-strand runs anti-parallel (one strand is 5 to 3 whilethe other strand is (3 to 5)
Very stable, due to very precise base-stacking and precise
hydrgogen bonding. Hydrogen bonding follows Chargaffsprinciple: G C and A T or (A + G) / (T + C) = 1
Each nucleotide is linked to one another by phosphodiester bond
Different types of structure of the DNA double helix:1. B-DNA 2. A-DNA 3. Z-DNA
a. Helix Right-handed Right-handed Left-handedb. # of basepairs per 360 0
turn
10 11 12
c. Height of each 360 0 turn
34 A 0 > 34 A 0 < 34 A 0
d. Breadth 20 A 0 < 20 A 0 > 20 A 0
DNA Supercoilingthe formation of additional coils in DNA due to twisting forces2 types:a. Negative supercoiling
> an underwound DNA molecule> have fewer right-hand turns than expected from thenumber of their base pairs
b. Positive supercoiling> an overwound DNA molecule> have more right-hand turns than expected from thenumber of their base pairs
DNA denaturation/DNA melting
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
5/29
ability of the DNA to separate into its two component strandsbrought about by high temperature (e.g. 100 0 C)
DNA renaturation/ DNA reannealingability of the complementary single-stranded DNA to formback the double helix
Types of DNA sequences:1. Highly repeated DNA sequencesa. satellite DNAsb. minisatellite DNAsc. microsatellite DNAs
2. Moderately Repeated DNA sequencesa. Repeated DNA sequences with coding functionsb. Repeated DNA sequences that lack coding functions
3. Non-repeated DNA sequences
Pseudogenes
sequences that are clearly homologous to functional genes,but have accumulated mutations that render themnonfunctional
TansposonsDNA segments capable of moving from one place in thegenome to another can cause mutation
B. ChromatinDNA molecules with associated histones and smaller amounts proteinsconsist of repeating units called nucleosomes
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
6/29
Histonesare heterogenous group of closely related arginine and lysinerich basic proteins (Arg and Lys help histones to bind tightlyto the negatively charged sugar phosphate backbone of DNA)provide for the compaction of chromatin
* Prokaryotic DNA is naked because it is not associated withhistones.
Types of Chomatin:1. Euchromatin
chromatin that is not highly compacted and maybetranscriptionally active
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
7/29
2. Heterochromatinchromatin that is highly compacted and is usuallytranscriptionally inactive2 types:a. Constitutive heterochromatin
> chromatin that remains in the compacted state in allcells at all times, thus, represents DNA that is permanentlysilenced
b. Facultative heterochromatin> chromatin that has been specifically inactivated during
certain phases of an organisms life
C. Chromosomesstructures made up of 2 coiled filaments which are productsof DNA replication during the S phasemade up of 2 chromatids (sister chromatids)Structure:a. centromere
> join the 2 sister chromatids> a boundary that separate the 2 arms> serve as handles, which allow mitotic spindles to attachto the chromosome during cell division
b. p short armc. q long armd. telomeres
> are hexameric DNA repeats (TTAGGG) found at the endsof chromosomes that serve to protect the chromosomesfrom degradation
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
8/29
Classification of chromosomes based on centromere location:
1. metacentric chromosomecentromere is centrally located
2. submetacentric chromosomecentromere is between the end and middle part
3. acrocentric chromosomecentromere is near the end
4. telocentric chromosomecentromere is at the end
Karyotypea photographic representation of all the chromosomes withina cellreveals how many chromosomes are found within an activelydividing somatic cell
Ploidy the number of chromosome copies within a cell1. Somatic cells
diploid number (46 or 23 pairs)
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
9/29
each nucleus has 2 copies of each chromosome (homologouschromosome), one from each paent
2. Germ cellshaplod number (22 autosomes and 1 sex chromosome)
Changes in chromosome number:1. Aneuploidy 1 extra or 1 less chromosome
Turner syndrome> inheriting 1 X chromosome with no corresponding X or Ychromosome
2. Polyploidy 3 or more of each type of chromosome
Klinefelter syndrome> inheriting 2 X chromosome and 1 Y chromosome
Downs syndrome (Trisomy 21)> 3 chromosome 21
Chromosomal aberrations and Human Disorders1. Inversions
a chromosome breaks in 2 places, and the segment betweenthe breaks become resealed into the chromosome in reverseorientation
2. Tanslocations
all or a piece of one chromosome becomes attached toanother chromosome
Philadelphia chromosome> a piece of chromosome 22 is taslocated to
chromosome 9> found in certain forms of leukemia
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
10/29
3. Deletionsa portion of chromosome is missing
Cri-du-chat syndrome> a portion of chromosome 5 is missing
4. Duplicationsa portion of chromosome is repeated
partial trisomy> a condition in which a number of genes are present in 3copies rather than 2 copies
II. Functional Organization of the Genome
In the 1860s, Gregor Mendel mated pea plants havingdifferent inheritable characteristic (height, seed color, seedshape, flower color, flower position, pod color, and pod shape) anddetermined the pattern by which these characteristics weretransmitted to offspring.
Genesbasic unit of inheritance that governs the character of aparticular traitin molecular terms, a segment of DNA containing theinformation for a single polypeptide or RNA moleculethere are 2 copies that control the development of each trait
Genotype particular genes an individual carriesPhenotype individuals observable traits
allelesalternate forms of agenecould be homozygous or heterozygous, dominant or recessive
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
11/29
homozygous dominant has a pair of dominant alleleshomozygous recessive has a pair of recessive allelesheterozygous has a pair of non-identical alleles
Mendels law of inheritance:1. Law of independent segregation
Even though the pair of alleles that governed a trait remainedtogether throughout life of an individual, they becomeseparated from one another during the formation of gametes
2. Law of independent assortmentThe segregation of the pair of alleles for one trait had no
effect on the segregation of alleles for another traitdoes not apply to all gene combination
Incomplete dominanceone allele of a pair is not fully dominant over its partner, so aheterozygous phenotype somewhere in between the 2homozygous phenotypes emergesEx: Red & White = Pink
CodominanceA pair of nonidentical alleles specify two phenotypes, whichare both expressed at the same time in heterozygotesEx: ABO blood types
PleiotrophyExpression of the alleles at just a single location on a
chromosome may have positive or negative effects on two or more traitsEx: Sickle cell anemia (HbS instead of HbA0
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
12/29
Inheritance patterns:1. Autosomal dominant
a pattern of inheritance in which the transmission of adominant allele on an autosome causes a trait to beexpressed
A aA AA (expressed) Aa (expressed)a Aa (expressed) aa (not expressed)
2. Autosomal recessivea pattern of inheritance in which the transmission of arecessive allele on both autosomes causes a trait to be
expressedA a
A AA (normal) Aa (carrier)a Aa (carrier) aa (affected)
3. X-linked recessivea mode of inheritance in which a mutation in a gene on the Xchromosome causes the phenotype to be expressed in males
(who are necessarily hemizygous for the gene mutationbecause they have only one X chromosome)
X YX XX (normal) XY (normal)X r XX r (carrier) X r Y(affected)
Non-Mendelian Inheritance:1. Maternal effect
Genotype of the mother directly determines the phenotypictraits of her offspring
2. Epigenetic inheritanceInheritance that is not encoded in the DNA sequence
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
13/29
The Human Genome Project (HGP)Started in 1990 and completed in 2003Project goals were to:a. Identify all the approximately 20,000-25,000 genes in
human DNAb. Determine the sequences of the 3 billion chemical base
pairs that make up the human DNAc. Store the information in databases (GenBank)d. Improve tools for data analysise. Transfer related technologies to the private sector f. Address the ethical, legal and social issues that may arise
from the project
CENTRAL DOGMA OF MOLECULAR BIOLOGY
ReplicationDuplication of the genetic material
TranscriptionFormation of a complementary RNA from a DNA template
Translation
Synthesis of proteins using the information encoded by anmRNA
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
14/29
DNA REPLICATION
Characteristics:1. Semiconservative with respect to parental strand
When DNA is replicated during the process of cell division (Sphase), each daughter duplex contains one strand from theparent structure
2. Bidirectional with multiple origins of replication. (Single originonly in the case of prokaryotes)
OriC specific site on E. coli chromosome where replicationbeginsORC(Origin Recognition Complex)
specific site on eukaryotic chromosome where replicationbegins
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
15/29
3. Primed by short stretches of RNA called primeraccomplished by primase, a type of RNA polymerase
4. Semidiscontinuous with respect to the synthesis of new DNAsynthesis is always in the 5 to 3 directionsynthesis is continuous in one strand (leading strand) whilediscontinuous in another strand (lagging strand)
Okazaki fragmentssmall segments of DNA that are rapidly linked to longer pieces to form the lagging strand
Replication forkthe points at which the pair of replicated segments cometogether and join the nonreplicated segmentscorresponds to a site where:1. the parental double helix is undergoing strand separation2. nucleotides are being incorporated into the newly
synthesized complementary strands
Enzymes Involved in DNA synthesis:1. DNA helicase
Responsible for unwinding the DNA double helix
2. Single-stranded binding protein (ssBP)Prevent premature annealing of the single-stranded DNA todouble stranded DNA
3. DNA Topoisomerase/ DNA gyraseIt relaxes the supercoiling of the DNA, thus helping in theunwinding of the helixPrevents overwinding of the DNA2 types:a. Topoisomerase I
> catalyzes breaks in only one DNA strand
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
16/29
b. Topoisomerase II> catalyzes breaks in both strands of the DNA
4. Primase initiate synthesis of the primer
5. DNA polymeraseMajor enzyme used in replicating DNAExtend an existing nucleic acid strand in 5 to 3 directionHave 3 to 5 exonuclease activity that allows them toremove nucleotides that are not part of the double helix5 to 3 exonuclease activity is important for removingprimers
Can also perform excision and repair of damaged DNA
Properties of Eukaryotic DNA polymerasePolymerase Location Nucleus Nucleus Mitochondri
aNucleus Nucleus
Replication Yes No Yes No YesRepair No Yes No Yes No
AssociatedFunctions:5-3polymerase
Yes Yes Yes Yes Yes
3-5exonuclease
No No Yes Yes Yes
5-3exonuclease
No No No No No
6. LigaseCatalyzes the sealing of nicks (breaks) remaining in theDNA after DNA polymerase fills the gap left by primase
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
17/29
Steps in DNA synthesis:1. Recognition of origin of replication by an initiator protein2. Unwinding of the DNA helix3. Formation of the RNA primer 4. Elongation of the new DNA strand5. Termination of the RNA primer 6. Covalent bonding of Okazaki fragments by DNA ligase7. Cutting of the replicated portion by DNA Topoisomerase
DNA damageCan result from both endogenous causes and exogenouscauses such as ionizing radiation, hydrocarbons, oxidative
free radicals and chemotherapeutic agents
DNA repair systems:1. Mismatch repair
Corrects mismatches of base pairing usually due tomistakes made by DNA polymerase during replication
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
18/29
2. Base excision repair Required to correct the spontaneous depurination andspontaneous deaminationDeamination of cytosine causes it to be converted to uracil
3. Nucleotide excision repair Remove UV light-induced DNA damage as well as DNAdamage from environmental chemicals which formpyrimidine-pyrimidine dimmers
4. Double-stranded DNA repair
Used to repair damage from ionizing radiation, oxidative freeradicals, or chemotherapeutic agents which causes DNAstrands to be severed2 types:a. Homologous recombination
> type of repair which takes advantage of sequenceinformation available from the unaffected homologouschromosome for proper repair of breaks
b. Non-homologous end joining> permits the joining of ends even if there is no sequencesimilarity between them
Xeroderma pigmentosumAn autosomal recessive genetic disorder of DNA repair inwhich patients carry mutations in the nucleotide repair
enzymes (defective repair of thymine dimers)Affected individuals are prone to develop multiple skincancers
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
19/29
TRANSCRIPTION
A process by which an RNA molecule is polymerized on aDNA template with the aid of various proteinsProduces all types of RNAs:1. rRNA/ ribosomal RNA
transcribed by RNA polymerase I80% of the total RNA in the cellAssociate with proteins to form ribosomes, which areimportant during protein synthesis as they containpeptidyl transferase activity
2. tRNA/ transfer RNAtranscribed by RNA polymerase IIsmallest of all the RNAsclover-leaf structureFunctions in protein synthesis:a. Carry the appropriate amino acidb. Provide a mechanism by which nucleotide
information can be translated to amino acidinformation through its anticodon
3. mRNA/ messenger RNAtranscribed by RNA polymerase IIlinear with hairpin loop structurecarries genetic information from DNA to cytosol for translation
4. Other types of RNA:a. Small nuclear RNAs (snRNAs)b. Small nucleolar RNAs (snoRNAs)c. Small interfering RNAs (siRNAs)d. micro RNAs (miRNAs)
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
20/29
Gene structure:
1. Exons coding region2. Introns noncoding region3. upstream sequences
noncoding regions at the 5 end to the 1 st exon4. Downstream sequences
found at the 3 end5. Promoters
are DNA conserved sequences that select or determine thestart site of RNA synthesisincludes:a. TATA box
Contains sequences of conserved nucleotides thatsignals start of transcriptionDirects RNA polymerase II to the correct site
b. CAAT box & GC boxSpecifies the frequency of initiating transcription
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
21/29
Steps in RNA transcription:1. Initiation
Binding of RNA polymerase to the promoter region
2. ElongationComplementary base pairing occur ( C G; A U)
3. TerminationA termination code in the DNA indicates where transcriptionwill stop
Pre-mRNA/ heterogenous nuclear RNA (hnRNA)
Large group of RNA molecules that found only in the nucleusand is usually altered by splicing and other modificationsbefore it exits the nucleusContains segments of transcribed introns
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
22/29
RNA processing reactions:1. Addition of a 5 cap
The 5 end of RNA is capped by a methyl guanosine residue,which protects it from degradation by 5 exonucleases duringelongation of the RNA chainAlso helps the transcript bind to the ribosome during proteinsynthesis
2. Addition of a poly (A) tail at the 3 end
3. Intron removal
Splice site sequences, which indicate the beginning (GU) andending (AG) of each intron, are found within the primary RNAtranscript
SpliceosomeSplice or join exons together to form the mature RNA
TRANSLATIONCodon
A sequence of three nucleotides in mRNA that functions intranslationStart codon: AUG (codes for methionine)Stop codon: UAA, UAG, UGA
Genetic code
Manner in which the nucleotide sequences of DNA encodethe information for making protein productsEach codon specifies a particular amino acid or the end of translationCharacteristics of the genetic code:1. Specificity
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
23/29
A particular codon always codes for the same aminoacid
2. UniversalitySpecificity has been conserved from very early stagesof evolution, with only slight differences in the manner in which the code is translated (* In the mitochondria,UGA codes for trp)
3. Redundancy/DegeneracyA given amino acid may have more than 1 tripletcoding for it
4. Nonoverlapping and commalessThe code is read from a fixed starting point as acontinuous sequence of bases, taken 3 at a time
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
24/29
Components required for translation:1. Amino acids
If one amino acid is missing, translation stops at thecodon specifying that amino acid
2. mRNA serves as template for synthesis of the polypeptide3. Aminoacyl-tRNA synthetase
Catalyzes a 2-step reaction that results in the covalentattachment of the carboxyl group of an amino acid to the3 end of its corresponding tRNA
4. Protein factors for initiation, elongation and termination5. ATP and GTP as source of energy
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
25/29
6. Ribosomes which are functionally competent2 subunits:a. Larger subunit
> catalyzes the formation of the peptide bonds thatlink amino acid residues in a protein
b. Smaller subunit> responsible for the accuracy of translation byensuring correct base-pairing between the codon inthe mRNA and the anticodon of the tRNA
3 binding sites for tRNA molecules:a.
A - binds an incoming aminoacyl-tRNA as directed bythe codon currently occupying the siteb. P - occupied by peptidyl-RNA, which carries the chain
of amino acids that has already been synthesizedc. E - occupied by the empty tRNA as it is about to exit
the ribosome
7. Transfer RNA
One specific type of tRNA is required per amino acidCarry a specific amino acid at its 3 endContains an anticodon
Anticodon> recognizes a specific codon on the mRNA> specifies the insertion into the growing polypeptide chainof the amino acid carried by that tRNA
> binding to the mRNA codon follows the rules of complementary and antiparallel binding, that is, the mRNAcodon is read 5-3 by an anticodon pairing in the flipped(3-5) orientation
Wobble hypothesis
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
26/29
> nontradional base-pairing between the 5 nucleotide (1 st
nucleotide) of the anticodon with the 3 nucleotide (lastnucleotide) of the mRNA codon> there need not be 61 tRNA species to read the 61 codonscoding for amino acids
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
27/29
Steps in Protein Translation:
1. InitiationAssembly of all the components of thetranslation system(ribosomes, mRNA, aminoacyl tRNA, GTP, initiator factors)
2. ElongationAddition of amino acids to carboxyl end of the growing chainRibosomes moves from the 5 end to the 3 end of the mRNARequires participation of GTP and elongation factorsFormation of peptide bonds is catalyze by peptidyltransferase activity of rRNA
3. TerminationRelease factor in eukaryote, eRF, recognizes all threetermination codons
Polysomes/ polyribosomeComplex of 1 mRNA and a number of ribosomes
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
28/29
Post-translational modification:1. Trimming2. Covalent modifications: phosphorylation, glycosylation,hydroxylation, carboxylation, farnesylation, acetylation
Consequences of altering the nucleotide sequence:1. Silent mutation
Codon containing the changed base may code for the sameamino acidEx: UCA UCU (still codes for serine)
2. Missense mutation
Codon containing the changed base may code for a differentamino acidEx: UCA (serine) CCA (proline)
3. Nonsense mutationCodon containing the changed base may become atermination codonEx: UCA (serine) UAA (stop codon)
4. Other mutations:a. Splice site mutations
Alter the way in which introns are removed from the pre-mRNA molecules thus producing aberrant proteinsEx: thalassemias
b. Frame- shift mutations
1 or 2 nucleotides are either deleted from or added to thecoding region of a message sequenceEx: Cystic fibrosis (due to a deletion of 3 nucleotides fromthe coding region of a gene, resulting in the loss of phenylalanine at the 508 th position)
-
8/7/2019 Part 11 lecture (eukaryotic nucleus)
29/29
c. trinucleotide repeat expansionA sequence of 3 bases that is repeated in tandem willbecome amplified in a number so that too many copies of thetriplet occur
Disease Repeatsequence
Normal # Diseasestate #
Location
1. Huntington CAG 11-34 42-100 Protein codingregion
2. Fragile X CGG 6-54 250-4000 5 untranslatedregion
3. Myotonic
dystrophy
CTG 5-30 >50 5 untranslated
region