Gilchrist et al. Supplementary Material Supplementary...
Transcript of Gilchrist et al. Supplementary Material Supplementary...

Gilchrist et al. Supplementary Material Supplementary Figure S1
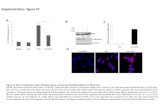
Supplementary Figure S1. Time Course of Gene Expression Changes in Response to NELF Depletion. (A) S2 cells were treated with dsRNA against NELF-B and -E (circles), mock-treated (triangles), or untreated (diamonds). Cell aliquots were harvested at 24, 40, 56, and 72 hours, and relative gene expression was determined by RT-PCR. The name of the gene analyzed is given above each graph, and data points represent mean of two independent samples assayed in duplicate (error bars depict range). Shown are the changes in the levels of transcripts encoding the NELF-E subunit, upregulated genes hsp70 and oaf, and downregulated genes syx4, CG11709, and CG5399. (B) S2 cells were treated with dsRNA against NELF-B and -E or mock treated, and cell aliquots were harvested at 24, 40, 56, and 72 hours. Whole cell extracts from equivalent culture volumes were separated by SDS-PAGE, blotted to nitrocellulose, and proteins were detected with the indicated antiserum. Mock-treated extracts diluted 1:10 and 1:3 were included to estimate the efficiency of depletions.
1

Supplementary Figure S2
Supplementary Figure S2. Validation of Microarray Results. To verify the changes in RNA levels observed in microarray experiments, RT-PCR was performed on independent samples to assess the effects of NELF-RNAi on 26 upregulated and 10 downregulated genes. RNA from independent samples that were untreated, mock-treated or NELF-depleted was reverse transcribed and individual RNA levels determined. Relative gene expression was calculated from microarray analysis or RT-PCR as described in Experimental Procedures. (A) Change in expression of representative upregulated genes in response to NELF depletion. Each bar represents the mean of three biological replicates +/- SEM. (B) downregulated genes were analyzed as in (A).
2

Supplementary Figure S3
Supplementary Figure S3. Pol II and NELF Are Associated with Gene Promoters. ChIP signals obtained for Pol II and NELF-E in untreated cells are shown for upregulated genes rho, net, CG10082, and CG6934 as well as NELF-unaffected genes beta-tubulin56D and lethal(3)04053 (CG11238). Gene positions given designate the center of each primer pair used with respect to the transcription start site for each gene. Error bars represent the SEM (n=3). (B) Permanganate genomic footprinting analysis on the downregulated genes CG5399, CG13117, syx4, CG30456, and the upregulated genes mfas and oaf. The locations of a subset of thymines are indicated at the right of each panel.
3

Supplementary Figure S4
Supplementary Figure S4. NELF-depletion does not change Pol II Serine-2 phosphorylation at NELF-Regulated Genes. ChIP signals obtained for Pol II (Rpb3, top panel) and Pol II phosphorylated at Serine-2 (Ser2-P, middle panel) were used to calculate the ratio of Rpb3 to Ser-2P (Rpb3/Ser2-P, lower panel) for mock-treated or NELF-depleted cells. Data are shown for NELF-unaffected genes RpL3 and eIF-5C, upregulated genes Hsp70 and oaf, and downregulated genes TepII and nocturnin. Gene positions given designate the center of each primer pair used with respect to the transcription start site for each gene. Error bars represent range of two biological replicates (n=2). Duplicate IPs were performed for each replicate.
4

Supplementary Figure S5
Supplementary Figure S5. NELF Depletion Alters Chromatin Organization at the Tl Gene. (A) Histone H3 ChIP signal the downregulated gene Tl in Mock-treated and NELF-depleted S2 cells. (B) Nucleosome positioning at Tl is altered by depletion of NELF. Chromatin from untreated, NELF-depleted, and mock-treated cells was digested with MNase. DNA corresponding to mono-nucleosomes (~150 bp) was used as a template for quantitative PCR with primer pairs spanning the Tl transcription start site. DNA sheared by sonication was used as a reference standard.
5

Supplementary Figure S6
Supplementary Figure S6. DNAse I Hypersensitivity changes in NELF-depleted cells. DNAse I hypersensitive sites were mapped for the promoter regions of the downregulated gene TepII and the NELF-unaffected gene Dr. Following 72 hour mock-treatment or NELF RNAi, nuclei were isolated from Drosophila S2 cells and incubated with increasing concentrations of DNAse I (1.25, 2.5, 5, and 10 units/ml). Digestions were stopped with EDTA, DNA was purified, and hypersensitive sites were visualized with ligation-mediated PCR. Transcription start sites are indicated by arrows.
6

Supplementary Figure S7
Supplementary Figure S7. NELF-depletion Leads to Aberrant Histone RNA Polyadenylation but Does Not Alter Histone Protein Levels. (A) Following 72 hour treatment with NELF-B and -E dsRNA, total RNA was harvested from S2 cells and reverse transcribed using either an oligo(dT) primer or random hexamers. The apparent change in gene expression detected with each primer type (relative to mock-treated cells) was quantified using RT-PCR. Values shown represent the mean of three biological replicates +/- SEM. (B) S2 cells were treated with dsRNA against NELF-B and -E or mock treated, and cell aliquots were harvested at 72 hours. Whole cell extracts from equivalent culture volumes were separated by SDS-PAGE, blotted to nitrocellulose, and proteins were detected with antiserum against NELF-E or Histone H4. Mock-treated extracts diluted 1:10 and 1:3 were included to estimate the efficiency of depletions.
7

Supplementary Table S1.
Complete List of 241 Transcripts Affected by NELF-depletion.
Nelf-dep. vs Untr.
Nelf-dep. vs Mock-tr.
Average
Affymetrix Probe ID
Gene Common Name
Fold Change
p-Value Fold Change
p-Value Fold Change
1636947_at CG9598-RA CG9598 6.73 3.91E-12 5.71 4.21E-11 6.22 1632298_s_at CG3359-RB mfas 3.54 0.00002 6.78 1.69E-17 5.16 1626821_s_at CG6489-RA Hsp70Bc 4.17 0.00001 5.43 6.40E-08 4.80 1632019_s_at CG9008-RA CG9008 4.21 4.42E-07 3.88 1.20E-06 4.05 1635712_at CG2247-RA CG2247 4.32 1.87E-33 3.66 3.60E-25 3.99 1632841_x_at CG6489-RA Hsp70Bc 3.75 0.00007 3.45 9.26E-06 3.60 1626867_at CG31613-RA HistoneH3 3.43 7.54E-12 3.74 2.40E-13 3.58 1639571_s_at CG18743-RA Hsp70Ab 3.25 2.12E-07 3.03 3.00E-07 3.14 1638308_s_at CG31613-RA HistoneH3 2.84 1.93E-35 2.94 4.57E-40 2.89 1634152_at CG12242-RA GstD5 3.00 3.20E-19 2.59 2.22E-21 2.80 1626218_s_at CG9884-RB oaf 2.66 2.77E-13 2.91 4.80E-15 2.79 1640802_at HDC19553 HDC19553 3.16 9.31E-08 2.39 0.00005 2.77 1640262_at CG30079-RA CG30079 2.91 0.00026 2.55 0.00053 2.73 1624913_s_at CG9884-RC oaf 2.59 4.60E-19 2.81 2.66E-18 2.70 1627494_s_at CG9884-RA oaf 2.70 0 2.61 0 2.65 1627446_at CG11450-RA netrin 3.11 2.97E-07 2.17 0.00013 2.64 1635044_at CG4183-RA Hsp26 2.83 3.79E-08 2.44 1.20E-10 2.63 1638432_a_at CG10082-RA CG10082 2.70 0.00003 2.52 5.87E-06 2.61 1635030_at CG12868-RA CG12868 2.75 3.65E-34 2.39 5.43E-27 2.57 1641413_s_at CG3074-RB CG3074 2.76 1.03E-07 2.26 2.92E-06 2.51 1635462_at CG1004-RA rhomboid 2.53 7.94E-14 2.11 1.69E-10 2.32 1629061_s_at CG32041-RB Hsp22 2.58 1.26E-12 1.98 2.06E-09 2.28 1623281_s_at CG17949-RA HistoneH2B 2.28 1.56E-22 2.20 6.02E-21 2.24 1624203_s_at CG3903-RA Gli 2.43 1.00E-10 1.99 2.37E-10 2.21 1629827_s_at CG31449-RA Hsp70Ba 2.24 0.00042 2.17 0.00065 2.20 1640932_s_at CG32626-RA CG32626 2.39 5.99E-09 1.98 1.65E-06 2.18 1639997_s_at CT39116 CT39116 2.14 1.86E-19 2.01 4.03E-15 2.07 1631994_a_at CG33048-RA Mocs1 2.07 0.00091 2.06 0.00095 2.06 1631321_s_at CR31614-RA Histone pseud. 1.95 4.69E-19 2.16 1.44E-14 2.06 1639704_at CG14695-RA CG14695 1.96 1.42E-11 2.14 6.43E-14 2.05 1626220_at CG3448-RB CG3448 2.11 2.61E-16 1.87 5.30E-11 1.99 1629740_at CG31617-RA HistoneH1 1.85 1.08E-16 2.07 1.81E-20 1.96 1635666_at CG17949-RA HistoneH2B 1.90 1.26E-44 1.97 2.80E-45 1.93 1631628_s_at CG11765-RA Prx2540 1.82 8.50E-15 2.04 9.48E-20 1.93 1641302_at CG6934-RA CG6934 1.79 7.42E-07 2.07 1.75E-07 1.93 1628353_at CG10045-RA GstD1 1.97 0 1.86 0 1.92 1640884_at CG15784-RA CG15784 2.09 2.54E-19 1.72 7.12E-09 1.91 1632345_at CG8353-RA CG8353 1.83 0.00012 1.96 6.63E-08 1.89 1635486_at CG33468-RA CG33468 2.17 8.28E-07 1.62 0.00099 1.89 1626061_at CG30078-RA CG30078 2.09 1.46E-08 1.67 0.00001 1.88
8

1635715_at CG31611-RA HistoneH4 1.89 3.50E-29 1.85 1.50E-18 1.87 1640912_s_at CG11066-RB CG11066 2.03 1.78E-38 1.70 4.97E-34 1.86 1640335_at CG31304-RA CG31304 1.78 7.70E-07 1.93 3.24E-08 1.85 1628683_at CG6272-RA CG6272 1.89 3.90E-41 1.76 1.11E-35 1.83 1634019_at CG2064-RA CG2064 1.79 0 1.86 0 1.82 1625672_s_at CT33405 CT33405 1.67 1.73E-11 1.92 9.31E-27 1.79 1636943_s_at CG18525-RA Spn5 1.65 6.78E-25 1.89 1.09E-21 1.77 1630964_at CG15211-RA CG15211 1.87 2.84E-14 1.66 6.40E-16 1.76 1632639_at CG13941-RA CG13941 1.79 9.77E-15 1.73 2.94E-12 1.76 1625759_at CG18801-RA Ku80 1.83 4.76E-07 1.68 0.00048 1.76 1625005_s_at CG1683-RB Ant2 1.80 2.92E-08 1.71 2.47E-07 1.75 1625925_at CG31618-RA HistoneH2A 1.70 5.32E-12 1.73 8.07E-09 1.72 1632958_a_at CG15675-RA CG15675 1.80 6.38E-06 1.60 0.00056 1.70 1637282_at CG6171-RA CG6171 1.56 0.00041 1.84 2.95E-07 1.70 1641367_at CG31685-RA CG31685 1.61 2.90E-08 1.76 4.44E-08 1.69 1623122_at CG17294-RA CG17294 1.68 7.01E-15 1.64 1.33E-10 1.66 1629681_x_at CG4068-RA CG4068 1.64 1.14E-12 1.62 2.69E-13 1.63 1633622_at CG3008-RA CG3008 1.67 2.56E-24 1.59 2.17E-16 1.63 1639694_s_at CG10102-RA CG10102 1.54 1.39E-09 1.69 8.23E-10 1.61 1630975_at CG2909-RA CG2909 1.63 1.57E-06 1.56 1.00E-05 1.60 1623227_s_at CG30147-RB CG30147 1.66 1.05E-11 1.49 4.69E-08 1.57 1626801_at CG10638-RA CG10638 1.59 2.08E-36 1.55 5.20E-23 1.57 1639054_s_at X59837 X59837 1.75 8.59E-19 1.38 1.52E-07 1.56 1639637_a_at CG3376-RB CG3376 1.47 0.00019 1.66 1.48E-06 1.56 1628402_a_at CG3881-RA CG3881 1.61 0.00003 1.51 0.00002 1.56 1636600_a_at CG6803-RA CG6803 1.59 4.16E-12 1.52 5.58E-19 1.56 1624800_at CG5224-RA CG5224 1.52 1.00E-08 1.56 1.90E-09 1.54 1627330_at CG10189-RA CG10189 1.51 2.02E-08 1.56 2.19E-09 1.53 1625744_at CG17530-RA CG17530 1.59 5.62E-29 1.47 3.65E-20 1.53 1623543_at CG5807-RA CG5807 1.61 4.34E-13 1.42 4.60E-08 1.52 1630934_at X51967 X51967 1.65 4.36E-23 1.38 4.24E-06 1.51 1637286_a_at CG12437-RB raw 1.69 9.19E-24 1.32 3.26E-07 1.51 1635684_a_at CG2999-RA unc-13 1.62 4.46E-20 1.39 2.26E-09 1.50 1635356_at CG8816-RA CG8816 -1.54 1.59E-07 -1.46 0.00004 -1.50 1638050_s_at CG3090-RA Sox14 -1.47 3.86E-07 -1.54 6.90E-09 -1.51 1637635_at CG4766-RA CG4766 -1.48 1.49E-14 -1.54 4.69E-19 -1.51 1623787_at CG7144-RA CG7144 -1.49 8.04E-06 -1.53 1.77E-06 -1.51 1630503_at CG6045-RA CG6045 -1.57 6.21E-06 -1.47 0.00017 -1.52 1627589_s_at CG9520-RB CG9520 -1.55 2.05E-20 -1.50 2.49E-19 -1.53 1630038_at CG3027-RA pyd3 -1.48 3.47E-16 -1.57 2.56E-21 -1.53 1638409_at CG12030-RA CG12030 -1.62 3.22E-26 -1.45 1.85E-25 -1.54 1623594_at CG1363-RA blow -1.58 0.00005 -1.56 0.00021 -1.57 1636193_at CG13893-RA CG13893 -1.56 1.16E-21 -1.59 1.04E-19 -1.57 1628974_at CG32711-RA CG32711 -1.67 6.13E-06 -1.48 0.0007 -1.57 1625613_at CG11899-RA CG11899 -1.57 8.63E-12 -1.58 3.55E-11 -1.58 1629468_at CG9372-RA CG9372 -1.72 5.11E-16 -1.44 5.57E-08 -1.58 1629205_at CG32032-RA CG32032 -1.63 5.95E-16 -1.56 4.66E-13 -1.59 1639575_at CG18446-RA CG18446 -1.57 1.63E-06 -1.63 6.90E-08 -1.60
9

1631121_at CG7267-RB CG7267 -1.65 2.84E-06 -1.56 0.00006 -1.60 1623813_at CG16887-RA CG33306 -1.74 1.13E-06 -1.49 0.00073 -1.62 1634619_at CG4026-RA CG4026 -1.73 1.46E-10 -1.54 0.00003 -1.63 1630375_at CG7635-RA CG7635 -1.76 7.81E-07 -1.55 0.00015 -1.66 1625257_s_at CG31523-RC CG31523 -1.63 9.01E-16 -1.72 1.94E-19 -1.67 1639443_at CG13631-RA CG13631 -1.73 2.06E-08 -1.66 0.00002 -1.69 1627795_at CG6747-RA Ir -1.64 3.18E-12 -1.75 2.46E-15 -1.70 1627073_a_at CG10126-RB CG10126 -1.67 1.10E-11 -1.74 2.20E-13 -1.70 1633353_s_at CG18408-RI rexin -1.63 3.65E-08 -1.79 2.68E-11 -1.71 1631568_a_at CG32717-RB sdt -1.67 0.00006 -1.75 2.26E-06 -1.71 1626857_at CG4408-RA CG4408 -1.77 0.00078 -1.65 0.00093 -1.71 1624617_at CG1665-RA CG1665 -1.79 2.18E-22 -1.64 1.05E-13 -1.72 1641164_s_at CG11163-RA CG11163 -1.81 7.41E-23 -1.66 2.78E-19 -1.74 1639671_at CG12919-RA eiger -1.90 5.06E-15 -1.58 5.24E-10 -1.74 1626434_s_at AE003485 AE003485 -1.68 5.77E-07 -1.81 1.28E-06 -1.75 1640918_at CG32145-RA ome -1.73 0.00028 -1.79 0.00011 -1.76 1623950_s_at CG2198-RB Ama -1.78 0.00022 -1.74 0.00002 -1.76 1624054_at CG5992-RA Adgf-A -1.86 3.82E-06 -1.66 0.00002 -1.76 1634074_a_at CG2913-RA yin -1.80 1.80E-38 -1.73 4.75E-33 -1.77 1641496_a_at CG5896-RA CG5896 -1.95 1.91E-18 -1.58 5.01E-09 -1.77 1623565_at CG17278-RA CG17278 -1.89 5.74E-25 -1.64 5.37E-17 -1.77 1623624_at CG14869-RA CG14869 -1.78 5.97E-06 -1.77 8.21E-06 -1.78 1632852_s_at CG2718-RB Gs1 -1.76 3.85E-06 -1.80 8.11E-07 -1.78 1627302_at CG3174-RA Fmo-2 -1.86 1.47E-32 -1.73 1.93E-25 -1.80 1640202_at CG4475-RA Idgf2 -1.89 1.75E-26 -1.74 7.24E-18 -1.82 1637983_s_at CG10365-RC CG10365 -1.85 3.05E-15 -1.81 3.87E-14 -1.83 1637430_s_at CG1572-RA CG1572 -1.87 4.73E-20 -1.81 3.54E-17 -1.84 1624971_at CG5254-RA CG5254 -1.92 2.11E-19 -1.77 9.66E-15 -1.84 1632790_at CG7863-RA dream -1.91 6.76E-06 -1.80 0.00005 -1.86 1634366_at CG32185-RA CG32185 -1.93 2.25E-12 -1.79 2.51E-07 -1.86 1638669_at CG11395-RA CG11395 -2.04 8.01E-13 -1.68 5.23E-07 -1.86 1634805_at CG14642-RA CG14642 -1.80 0.00008 -1.96 4.01E-06 -1.88 1623643_s_at CG4559-RA Idgf3 -1.94 4.79E-40 -1.84 3.91E-32 -1.89 1624760_at CG8306-RA CG8306 -2.02 3.54E-10 -1.76 3.40E-08 -1.89 1635933_at CG4496-RA CG4496 -1.94 3.93E-06 -1.86 0.00007 -1.90 1631486_at CG33338-RA p38c -2.07 1.28E-07 -1.77 0.00007 -1.92 1641598_at CG8805-RA wun2 -2.03 3.94E-07 -1.81 2.50E-06 -1.92 1636815_a_at CG3779-RB numb -1.84 2.93E-09 -2.00 6.14E-12 -1.92 1641504_s_at CG3413-RB wdp -1.86 1.89E-07 -1.99 1.65E-11 -1.92 1633488_at CG3705-RA aay -2.02 5.13E-06 -1.85 0.00004 -1.93 1624720_s_at CG6043-RB CG6043 -1.97 0.00005 -1.90 0.00012 -1.94 1627060_s_at CG1106-RB Gel -2.04 1.03E-33 -1.84 1.10E-14 -1.94 1628947_s_at CG3297-RB mnd -1.91 1.24E-10 -2.00 4.36E-12 -1.95 1637370_at CG18550-RA yellow-f -2.06 1.22E-26 -1.86 1.47E-17 -1.96 1637645_at CG18321-RA miple2 -1.92 4.49E-13 -2.04 1.76E-13 -1.98 1639321_s_at CG5490-RB Tl -1.96 3.55E-06 -2.04 7.26E-07 -2.00 1641623_at CG15917-RA CG15917 -1.94 0 -2.08 0 -2.01 1623689_a_at CG14642-RA CG14642 -2.17 5.07E-15 -1.90 3.44E-10 -2.03
10

1640112_at CG8050-RA Cys -2.01 7.34E-07 -2.07 0.00008 -2.04 1633795_a_at CG17124-RA CG17124 -2.06 0.00004 -2.06 0.00004 -2.06 1626536_at CG6776-RA CG6776 -2.18 6.63E-22 -1.95 7.80E-16 -2.07 1639535_at CG3254-RA CG3254 -1.86 1.89E-20 -2.33 0 -2.09 1625737_at CG12133-RA CG12133 -2.34 6.54E-07 -1.86 0.00054 -2.10 1635370_at CG12840-RA Tsp42El -2.06 1.56E-07 -2.18 1.01E-08 -2.12 1625563_s_at CG14253-RB CG14253 -2.42 2.46E-13 -1.88 2.06E-08 -2.15 1630968_at CG13907-RA CG13907 -2.07 0.00001 -2.23 8.85E-07 -2.15 1628144_at CG14273-RA CG14273 -2.21 0 -2.11 1.05E-43 -2.16 1640957_at CG1787-RA Hexo2 -2.27 0 -2.07 0 -2.17 1634958_at CG9455-RA CG9455 -2.40 4.66E-17 -1.94 1.17E-09 -2.17 1636693_at CG4330-RA CG4330 -2.49 1.20E-09 -1.94 0.00003 -2.22 1640703_at CG30460-RC CG30460 -2.26 0.00015 -2.18 0.00034 -2.22 1625885_at CG4869-RA βTub97EF -2.33 2.14E-08 -2.14 7.27E-07 -2.23 1624957_a_at CG4821-RD Tequila -2.45 8.88E-08 -2.02 0.00006 -2.23 1625048_at CG8318-RA Nf1 -2.26 4.97E-08 -2.22 1.11E-07 -2.24 1635109_at CG5888-RA CG5888 -2.39 1.95E-23 -2.12 4.45E-17 -2.25 1641429_at CG17523-RA CG17523 -2.39 0.00005 -2.16 0.0005 -2.27 1638462_at CG18088-RA CG18088 -2.66 9.19E-11 -1.91 0.00001 -2.28 1633545_at CG7496-RA PGRP-SD -2.44 9.63E-20 -2.15 4.61E-12 -2.29 1626955_at CG12014-RA CG12014 -2.45 3.54E-09 -2.14 1.27E-06 -2.29 1638953_a_at CG2086-RA drpr -2.41 2.25E-09 -2.18 1.87E-10 -2.30 1624836_at CG13795-RA CG13795 -2.40 1.66E-16 -2.21 2.00E-13 -2.31 1626414_at CG15101-RA Jheh1 -2.58 6.59E-11 -2.04 1.06E-06 -2.31 1628465_a_at CG3960-RC CG3960 -2.34 6.86E-19 -2.34 6.12E-19 -2.34 1641065_s_at CG3153-RB CG3153 -2.21 1.41E-08 -2.48 3.77E-09 -2.34 1627287_at CG5966-RA CG5966 -2.17 0.00059 -2.54 0.00008 -2.36 1632964_at CG15678-RA CG15678 -2.59 6.73E-35 -2.17 2.50E-14 -2.38 1629572_a_at CG6953-RA fat-spondin -2.63 0 -2.15 8.04E-30 -2.39 1629889_s_at CG1803-RA regucalcin -2.47 1.23E-26 -2.33 5.66E-16 -2.40 1635691_at CG11491-RF br -2.26 0.00053 -2.54 0.00004 -2.40 1623388_at CG15861-RA CG15861 -2.50 0.00003 -2.33 0.00017 -2.41 1634468_at CG13397-RA CG13397 -2.49 7.64E-11 -2.36 1.33E-09 -2.43 1635930_at CG5264-RA btn -2.54 1.36E-12 -2.32 7.80E-11 -2.43 1639255_s_at CG1516-RI CG1516 -2.40 7.07E-34 -2.48 3.54E-39 -2.44 1625369_at CG10877-RA CG10877 -2.81 2.11E-07 -2.08 0.00046 -2.45 1641428_at CG3616-RA Cyp9c1 -2.47 2.41E-22 -2.43 1.68E-21 -2.45 1637269_at CG6310-RA CG6310 -2.60 4.13E-34 -2.31 9.48E-22 -2.46 1627277_s_at CG2930-RA CG2930 -2.48 2.56E-36 -2.49 1.60E-36 -2.48 1636103_a_at CG31299-RB nocturnin -2.65 2.65E-09 -2.31 9.19E-09 -2.48 1632011_at CG31447-RA MESK4 -2.53 7.04E-29 -2.45 8.15E-27 -2.49 1635639_a_at CG32180-RA Eip74EF -2.40 6.92E-09 -2.64 4.92E-11 -2.52 1634396_at CG3303-RA CG3303 -2.75 3.00E-19 -2.30 1.52E-12 -2.52 1641157_at CG1888-RA CG1888 -2.40 8.36E-06 -2.66 9.75E-09 -2.53 1635470_at CG5008-RA GNBP3 -2.88 6.35E-18 -2.21 1.16E-09 -2.55 1629674_s_at CG10999-RA CG10999 -2.55 1.55E-17 -2.59 2.71E-18 -2.57 1627000_s_at CG6231-RC CG6231 -2.45 8.55E-15 -2.69 2.28E-18 -2.57 1627872_at CG3770-RA CG3770 -2.62 6.75E-33 -2.54 1.09E-30 -2.58
11

1640075_a_at CG3424-RA CG3424 -2.48 3.75E-09 -2.70 4.37E-11 -2.59 1629375_at CG11400-RA CG11400 -3.08 6.61E-29 -2.15 5.77E-17 -2.62 1633975_s_at CG12214-RB CG12214 -2.79 0.00006 -2.48 0.00097 -2.64 1636736_s_at CG31274-RA CG31274 -2.89 2.68E-23 -2.47 3.62E-11 -2.68 1623494_at CG2723-RA ImpE3 -2.68 6.12E-08 -2.68 5.13E-08 -2.68 1630067_a_at CG7052-RA TepII -2.86 1.20E-18 -2.53 3.85E-14 -2.69 1639069_at CG18240-RA CG18240 -2.76 0 -2.71 0 -2.74 1628493_at CG6173-RA kal-1 -2.59 8.55E-11 -2.94 3.09E-14 -2.77 1636293_at CG2217-RA CG2217 -2.75 4.37E-06 -2.81 2.21E-06 -2.78 1632070_at CG4414-RA Ugt58Fa -3.34 3.14E-19 -2.30 1.30E-08 -2.82 1634983_s_at CG32521-RA CG32521 -2.87 9.31E-24 -2.83 1.68E-30 -2.85 1641192_at CG14523-RA CG14523 -3.08 2.50E-14 -2.63 2.63E-10 -2.86 1626530_at CG1878-RA CecB -3.26 4.78E-16 -2.49 4.46E-09 -2.88 1631179_s_at CG6680-RA CG6680 -3.16 2.27E-12 -2.66 5.31E-09 -2.91 1632313_at CG1998-RA CG1998 -3.30 0 -2.70 3.07E-39 -3.00 1623327_at S.C2R001709 S.C2R001709 -2.97 5.61E-45 -3.04 0 -3.01 1625249_at CG12664-RB ld14 -3.29 7.96E-12 -2.74 8.64E-08 -3.01 1639834_at CG14291-RA CG14291 -3.24 0 -2.80 0 -3.02 1631426_at CG32412-RA CG32412 -3.09 0 -2.95 0 -3.02 1625023_a_at CG11822-RA nAcRbeta-21C -3.63 7.21E-08 -2.47 0.00025 -3.05 1629430_s_at HDC18536 HDC18536 -3.41 3.74E-07 -2.72 0.00011 -3.07 1632299_at CG16827-RA alphaPS4 -3.06 0.00053 -3.19 0.00027 -3.13 1625114_at CG10391-RA Cyp310a1 -3.55 6.93E-09 -2.77 0.00001 -3.16 1627551_s_at CG18372-RA AttB -3.69 1.00E-27 -2.64 1.63E-14 -3.17 1625338_at CG16761-RA Cyp4d20 -3.13 5.74E-08 -3.21 2.19E-08 -3.17 1629442_at S.C2R000679 S.C2R000679 -3.80 7.58E-09 -2.55 0.00028 -3.17 1632490_at CG8008-RA CG8008 -3.10 3.60E-10 -3.26 6.36E-16 -3.18 1634978_at CG13117-RA CG13117 -3.39 2.35E-37 -3.07 2.40E-31 -3.23 1630065_at CG6912-RA CG6912 -3.48 1.10E-14 -2.99 1.18E-11 -3.24 1628581_at CG3884-RB CG3884 -3.83 6.04E-09 -2.71 0.00078 -3.27 1633701_at CG16985-RA CG16985 -3.58 1.55E-28 -3.09 1.72E-15 -3.33 1623299_at CG1794-RA Mmp2 -4.01 2.51E-08 -2.70 0.00034 -3.35 1632600_at HDC05924 HDC05924 -3.80 5.93E-08 -2.93 1.28E-06 -3.37 1624468_a_at CG10026-RC CG10026 -4.25 4.68E-09 -2.51 6.84E-06 -3.38 1625687_at CG10275-RA CG10275 -3.08 0.00025 -3.69 7.34E-06 -3.39 1627352_at CG8782-RA CG8782 -3.93 0.00005 -2.85 0.00013 -3.39 1639643_at CG18557-RA CG18557 -3.58 4.35E-07 -3.25 9.98E-06 -3.42 1633401_s_at CG30489-RA Cyp12d1-p -3.81 2.11E-34 -3.16 1.35E-32 -3.48 1622908_a_at CG10962-RA CG10962 -3.71 1.25E-25 -3.34 4.82E-21 -3.53 1625388_at CG4893-RA CG4893 -3.62 0.00006 -3.48 0.00006 -3.55 1629776_a_at CG6643-RA CG6643 -3.68 8.01E-09 -3.52 3.69E-08 -3.60 1640747_s_at CG8547-RA CG8547 -3.46 2.56E-07 -3.77 9.74E-10 -3.62 1630487_s_at CG32041-RA CG32041 -3.92 9.48E-11 -3.54 8.22E-07 -3.73 1623777_s_at CG1358-RA CG1358 -4.00 1.23E-22 -3.61 2.92E-16 -3.81 1629786_a_at CG31116-RD CG31116 -3.62 9.84E-10 -4.42 4.40E-14 -4.02 1628935_at CG10962-RB CG10962 -4.09 4.37E-07 -4.06 1.76E-08 -4.08 1636804_at CG14629-RA CG14629 -5.01 1.14E-11 -3.20 4.46E-06 -4.10 1629857_at CG10359-RA CG10359 -4.88 1.36E-06 -3.50 0.00057 -4.19
12

1636145_at CG7219-RA CG7219 -4.71 0.00002 -3.88 0.00048 -4.29 1627744_at CG15209-RA CG15209 -4.93 1.29E-34 -3.66 1.50E-27 -4.30 1627662_at CG6644-RA Ugt35a -4.62 4.09E-11 -4.22 1.41E-09 -4.42 1639257_s_at CG11739-RC CG11739 -5.42 1.66E-09 -3.89 8.44E-06 -4.65 1638808_at CG2715-RA Syx4 -5.25 8.79E-19 -5.00 2.39E-17 -5.13 1628884_at CG11709-RA PGRP-SA -5.87 0 -4.97 0 -5.42 1625227_at CG30456-RA CG30456 -5.87 2.72E-23 -5.00 5.20E-18 -5.43 1627746_at CG6124-RA CG6124 -5.95 3.73E-06 -4.96 0.00004 -5.45 1630233_at CG5399-RA CG5399 -6.25 0 -4.70 1.41E-34 -5.48 1629337_at CG32721-RA NELF-B -6.54 1.17E-10 -6.73 2.69E-15 -6.63 1639102_at CG5994-RA NELF-E -13.44 0 -12.08 0 -12.76
Supplementary Table S1. Complete List of 241 Transcripts Affected by NELF-depletion, and the two NELF subunits targeted by NELF RNAi. The fold-change and P value for each RNA species (including RNAi targets NELF-B and NELF-E) is given for NELF-depleted cells versus both Untreated and mock-treated samples. Microarray experiments were performed using Drosophila Genome 2.0 Genechip® arrays (Affymetrix, Santa Clara, CA) as described below. Starting with 1 μg of total RNA, biotin-labeled cRNA was produced using the Affymetrix 3’ Amplification One-Cycle Target labeling kit according to manufacturer’s protocol. For each array, 10 μg of amplified cRNAs were fragmented and hybridized to the array for 16 hours in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Slides were stained and washed as indicated in the Antibody Amplification Stain for Eukaryotic Targets protocol using the Affymetrix Fluidics Station FS450. Arrays were scanned with an Affymetrix Scanner 3000 and data obtained using the Genechip® Operating Software (Version 1.2.0.037). The resulting files were imported into the Rosetta Resolver system (Version 5.1), which performs data pre-processing and error modeling as described in {Weng, 2006 #1208}. Ratios were built and pairwise comparisons performed using the Rosetta Resolver system. RNA transcripts that were verified in independent RT-PCR experiments are shown in bold. ChIP analyses were performed on each of the genes shown in green (n= 16 downregulated genes, n=9 upregulated genes) for the data shown in Figures 5B and 6B, 6D.
13

14
Supplementary Table S2. The top 5 hexamers enriched in the -250bp to +50bp sequences, compared between the up-regulated and down-regulated genes
hexamer name Enriched in p-value
tataaa TATA box up-regulated genes 1.69e-05 gcgaga unknown down-regulated genes 2.81e-05 tatcga unknown up-regulated genes 2.13e-04 taaata TBP (yeast) up-regulated genes 2.76e-04 tcacaa unknown up-regulated genes 4.15e-04
Computational Analyses For each gene, we downloaded the -250bp to +50bp sequence from UCSC genome browser (build dm3) (http://genome.ucsc.edu/). We used a relatively large region for analysis as we are not certain that the annotations of the transcription start sites are completely accurate. Forty seven and one-hundred and seven sequences were obtained for the up- and down-regulated genes, respectively. We enumerated all 4096 possible hexamers. For each hexamer, we counted the number of sequences of up- and down-regulated genes containing at least one such hexamer in the -250bp to +50bp region. For each hexamer, a 2×2 contingency table was constructed with up- and down-regulated genes being the two categories and zero and greater than zero counts being the rows. The hexamers were then ranked in increasing order according to the p-value for the Fisher’s exact test on the resulting 2×2 contingency tables. The top 5 hexamers are listed above (Supplementary Table S1). It can be seen that tataaa is the most enriched hexamer in the upregulated genes compared to the downregulated genes. We also carried out the same computational analysis on a smaller region (-100bp to +50bp) of the same sequences. Three (tataaa, tatcga, and tcacaa) of the previous top five hexamers remained in the five most significant hexamers with tataaa (p-value=7.3e-05) being the top one. To account for possible variations in TATA box, we scanned for putative TATA box sequences in the -250bp to +50bp sequences of both up- and down-regulated genes using the TATA box position weight matrix (PWM) (id: M00252) from the TRANSFAC database (Knuppel et al., 1994). Using a PWM score cutoff that resulted in approximately 25% of all Drosophila genes containing a TATA box in the same region (-250bp to +50bp), 20 of the 47 (42.6%) upregulated genes contained at least one predicted TATA box whereas 18 of the 106 (17.0%) downregulated genes contained at least one predicted TATA box. This result suggests that the upregulated genes may be TATA box enriched compared to the downregulated genes (p-value=0.0011, Fisher’s exact test). It also suggest that the up-regulated genes may be TATA box enriched and the down-regulated genes TATA box depleted compared to promoters genome-wide. References Knuppel,R. et al. (1994) TRANSFAC retrieval program: a network model database of eukaryotic
transcription regulating sequences and proteins. J. Comput. Biol., 1, 191-198.