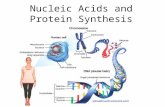
Nucleic Acids and Protein Synthesis. What are nucleic acids?
06 May 2008 Nucleic Acid Metabolism Andy Howard Introductory Biochemistry 6 May 2008.
-
Upload
allan-harmon -
Category
Documents
-
view
222 -
download
0
Transcript of 06 May 2008 Nucleic Acid Metabolism Andy Howard Introductory Biochemistry 6 May 2008.

06 May 2008Nucleic Acid Metabolism
Nucleic Acid Metabolism
Andy HowardIntroductory Biochemistry
6 May 2008

06 May 2008Nucleic Acid Metabolism p.2 of 56
What we’ll discuss Pyrimidine synthesis
PRPP Pathway to UMP Regulation Pathway to CTP
Purine synthesis IMP AMP, XMP, GMP Regulation
Reduction of riboNucs to deoxyNucs
dUMP to dTMP Salvage pathways Pyrimidine
catabolism Purine catabolism

06 May 2008Nucleic Acid Metabolism p.3 of 56
PRPP synthetase
Activation of ribose-5-P (see Calvin cycle, etc.) by ATP:-ribose-5-P + ATP PRPP + AMP
Has roles in other systems too
Phosphoribosyl pyrophosphate
PRPP synthetasePDB 2H06
215 kDa hexamerdimer shown; human

06 May 2008Nucleic Acid Metabolism p.4 of 56
Pyrimidine synthesis:carbamoyl aspartate
Uridine is based on orotate,which is derivated fromcarbamoyl aspartate
We’ve already seen the carbamoyl phosphate synthesis back in chapter 17 via carbamoyl phosphate synthetase
Carbamoyl phosphate + aspartate carbamoyl aspartate + Pi
via aspartate transcarbamoylase
Carbamoyl aspartate
Carbamoyl phosphate

06 May 2008Nucleic Acid Metabolism p.5 of 56
Aspartate transcarbamoylase
ATCase is the classic allosteric enzyme
E.coli version is inhibited by pyrimidine nucleotides and activated by ATP
CTP by itself is 50% inhibitory; CTP+ UTP is almost totally inhibitory
ATCasePDB 1D09Trimer of heterotetramers1 heterotetramer shown (cf. fig.18.11)E.coli

06 May 2008Nucleic Acid Metabolism p.6 of 56
Carbamoyl aspartate to dihydroorotate
Carbamoyl aspartate dehydrates and cyclizes to L-dihydroorotate via dihydroorotase
TIM barrel protein
Dihydro-orotate
PDB 1XGE76 kDa dimerE.coli

06 May 2008Nucleic Acid Metabolism p.7 of 56
Dihydroorotate to orotate
Ubiquinone acts as oxidizing agent reducing the 5 & 6 Carbons via dihydroorotate dehydrogenase
Some versions incorporate FMN
PDB 2E6F69 kDa dimerTrypanosoma cruzi

06 May 2008Nucleic Acid Metabolism p.8 of 56
Adding phosphoribose
Orotate + PRPP orotidine 5’-monophosphate + PPi
Usual argument re pyrophosphate hydrolysis
Enzyme: orotidine phosphoribosyl transferase Orotidine 5’-
monophosphate
PDB 2PS150 kDa dimerYeast

06 May 2008Nucleic Acid Metabolism p.9 of 56
Decarboxylation OMP
decarboxylated to form UMP via OMP decarboxylase
Bacterial forms are TIM barrel proteins
Acceleration is 1017-fold relative to uncatalyzed rate
PDB 1KLY54 kDa dimerMethanobacteriumthermoautotrophicum

06 May 2008Nucleic Acid Metabolism p.10 of 56
Eukaryotic variation Orotate produced in the
mitochondrion moves to the cytosol
UMP synthase combines the last two reactions—orotidine to OMP to UMP
OMP decarboxylasedomainof UMP synthasePDB 2P1F64 kDa dimerhuman

06 May 2008Nucleic Acid Metabolism p.11 of 56
UMP to UTP Uridylate kinase converts
UMP to UDP:UMP + ATP UDP + ADPenzyme is related to several amino acid kinases
Nucleoside diphosphate kinase exchanges di for tri:UDP + ATP UTP +ADP(non-specific enzyme)
Uridylate kinasePDB 2A1F163 kDa hexamerHaemophilusinfluenzae

06 May 2008Nucleic Acid Metabolism p.12 of 56
CTP synthetase
UTP + gln + ATP CTP + glu + ADP + Pi
Glutamine side-chain is amine donor
ATP provides energy sandwich (Rossmann) Enzyme is inhibited by CTP In E.coli, it’s activated by
GTP (makes sense!)
PDB 1S1M240 kDa tetramerdimer shownE.coli

06 May 2008Nucleic Acid Metabolism p.13 of 56
Purine synthesis Considerably more complex than
pyrimidine synthesis More atoms to condense and two rings to
make More ATP to sacrifice during synthesis Several synthetase (ligase) reactions
require ATP Based on PRPP, gln, 10-formyl THF, asp

06 May 2008Nucleic Acid Metabolism p.14 of 56
PRPP + gln to phospho-ribosylamine
PRPP aminated:PRPP + gln glu + PPi +5-phospho--D-ribosylaminevia glutamine-PRPP amidotransferase
transferase structure Product is unstable
(lasts seconds!)
1
PDB 1ECF120 kDa tetramerdimer shownE.coli

06 May 2008Nucleic Acid Metabolism p.15 of 56
Phospho-ribosylamine to GAR
Amine condenses with glycine to form glycinamide ribonucleotide (GAR)
ATP hydrolysis drives GAR synthetase reaction to the right PDB 2YRX
50 kDa monomerGeobacillus kaustophilus
2

06 May 2008Nucleic Acid Metabolism p.16 of 56
Formylation of GAR
10-formyl THF donates a formyl (-CH=O) group to end nitrogen with the help of GAR transformylase to form formylglycinamide ribonucleotide (FGAR)
Rossmann
FGAR
PDB 1MEO47 kDa dimer
human
3

06 May 2008Nucleic Acid Metabolism p.17 of 56
FGAR to FGAM
Glutamine sidechain is source of N for C=O exchanging to C=NH via FGAM synthetase to form formylglycinamidine ribonucleotide (FGAM):FGAR + gln + ATP + H2O FGAM + glu + ADP + Pi
PurS component ofFGAM synthetasePDB 1GTD37.4 kDa tetramerdimer shown
Methanobacterium
FGAM
4

06 May 2008Nucleic Acid Metabolism p.18 of 56
FGAM to AIR Cyclize FGAM to
aminoimidazole ribonucleotide
ATP drives the AIR synthetase reaction:
FGAM + ATP AIR + H2O + ADP + Pi
E.C. in Wikipedia is wrong:it should be 6.3.3.1
PDB 2V9Y147 kDa tetramerdimer shownhuman
Aminoimidazoleribonucleotide
5

06 May 2008Nucleic Acid Metabolism p.19 of 56
AIR to CAIR AIR is carboxylated;
expenditure of an ATP:AIR + HCO3
- + ATP carboxyaminoimidazole ribonucleotide + ADP + Pi + 2H+
AIR carboxylase E.coli version is two
enzymes; eukaryotes have a single enzyme
No cofactors!
PDB 2NSH149 kDa octamermonomer shownE.coli
CAIR
6

06 May 2008Nucleic Acid Metabolism p.20 of 56
CAIR+asp to SAICAR
CAIR + asp + ATP aminoimidazole succinylocarboxamide ribonucleotide + ADP + Pi
Enzyme is SAICAR synthetase
Domain 1: homolog ofphosphorylasekinase
Domain 2: ATP-bindingPDB 2CNQ34 kDamonomeryeast
7

06 May 2008Nucleic Acid Metabolism p.21 of 56
SAICAR to AICAR SAICAR aminoimidazole
carboxamide ribonucleotide + fumarate
Enzyme is adenylosuccinate lyase
Net result of two reactions is just replacing acid with amide;
That’s like first 2 reactions in urea cycle, except ADP, not AMP, is the product PDB 2PTR
203 kDa tetramerdimer shown; E.coli
8

06 May 2008Nucleic Acid Metabolism p.22 of 56
AICAR to FAICAR 10-formylTHF donates HC=O:
AICAR + 10-formylTHF formamidoimidazole carboxamide ribonucleotide + THF
Enzyme: AICAR transformylase Like step 3 Generally a bifunctional enzyme
combined with next step This part is like cytidine
deaminase (see below)
9
PDB 1THZ130 kDa dimer
chicken

06 May 2008Nucleic Acid Metabolism p.23 of 56
FAICAR to IMP We made it:
FAICAR inosine 5’-monosphosphate + H2O
Bifunctional enzyme; this part is called IMP cyclohydrolase or inosicase
Hydrolase part is like methylglyoxal synthase PDB 1PL0
260 kDa tetramerdimer shown; human
10

06 May 2008Nucleic Acid Metabolism p.24 of 56
So now we have a purine. What next?
Enzymatic conversions to AMP or GMP;Details on next few slides
AMP and GMP can be further phosphorylated to make ADP, GDP with specific kinases (adenylate kinase and guanylate kinase)
GTP made with broad-spectrum nucleoside diphosphate kinase

06 May 2008Nucleic Acid Metabolism p.25 of 56
IMP to adenylosuccinate
IMP + aspartate + GTP adenylosuccinate + GDP + Pi
Enzyme is adenylosuccinate synthase
Similar to step 7 in IMP synthesis
PDB 2V40101 kDa dimermonomer shownhuman

06 May 2008Nucleic Acid Metabolism p.26 of 56
Adenylosuccinate to AMP
Adenylosuccinate AMP + fumarate
Like reaction 8 in the IMP pathway; in fact, it uses the same enzyme, adenylosuccinate lyase
PDB 2PTR203 kDa tetramerdimer shown; E.coli

06 May 2008Nucleic Acid Metabolism p.27 of 56
IMP to XMP
IMP + H2O + NAD+ Xanthosine monophosphate + NADH + H+
Enzyme: IMP dehydrogenase
TIM-barrel, aldolase-like protein
PDB 1ME8221 kDa tetramer;monomer shownTritrichomonas foetus

06 May 2008Nucleic Acid Metabolism p.28 of 56
XMP to GMP
XMP + gln + H2O + ATP GMP + glu + AMP + PPi
Enzyme: GMP synthetase
Typical 3-layer sandwich
PDB 2DPL68 kDa dimerPyrococcus horikoshii

06 May 2008Nucleic Acid Metabolism p.29 of 56
Adenylate kinase
Reminder:ATP + AMP 2 ADP
Metal ions play a role in enzyme structure
Enzymes like this need to shield their active sites from water to avoid pointless hydrolysis of ATP
PDB 1ZIN24 kDa monomerBacillus stearothermophilus

06 May 2008Nucleic Acid Metabolism p.30 of 56
Guanylate kinase GMP + ATP GDP + ADP “P-loop”-containing ATP-
binding proteins Rossmann fold
PDB 2QOR22 kDa monomerPlasmodium vivax

06 May 2008Nucleic Acid Metabolism p.31 of 56
Purine control I: IMP level
Note that GTP is a cosubstrate in making AMP from IMP
ATP is a cosubstrate in making GMP from IMP
So this helps balance the 2 products

06 May 2008Nucleic Acid Metabolism p.32 of 56
Purine control II:feedback inhibition
PRPP synthetase inhibited by purines, but only at unrealistic concentrations of [Pur]
Step 1 (gln-PRPP amidotransferase) is allosterically inhibited by IMP, AMP, GMP
Adenylosuccinate synthetase is inhibited by AMP
XMP and GMP inhibit IMP dehydrogenase

06 May 2008Nucleic Acid Metabolism p.33 of 56
Making deoxyribonucleotides
Conversions of nucleotides to deoxynucleotides occurs at the diphosphate level
Reichard showed that most organisms have a single ribonucleotide reductase that converts ADP, GDP, CDP, UDP to dADP, dGDP, dCDP, and dUDP
NADPH is the reducing agent

06 May 2008Nucleic Acid Metabolism p.34 of 56
Ribonucleotide reductase heterotetramer
2 RNR1 subunits; each has a helical 220-aa domain 10-strand 480-aa structure
(thiols here) 5-strand 70-aa structure
2 RNR2 subunits; each has A diferric ion center A stable tyrosyl free radical
RNR1PDB 1R1R
258 kDa dimerE.coli
RNR2PDB 1PJ0
82 kDa dimerE.coli

06 May 2008Nucleic Acid Metabolism p.35 of 56
Mechanism of RNR (box 18.3)
Y122 in RNR2 is converted to stable free radical
Radical transmitted to RNR1 cys439 Cys439 reacts with substrate 3’-OH to form
free radical at C3’ Substrate dehydrates to carbonyl at C3’ and
free radical at C2’; S- formed at Cys462 Disulfide formed between Cys462,Cys225;
radical regenerated at Cys439

06 May 2008Nucleic Acid Metabolism p.36 of 56
Ribonucleotide reductase: control
ATP, dATP, dTTP, and dGTP act as allosteric modulators by binding to two regulatory sites on the enzyme
Activity site (A) regulates activity of catalytic site When ATP binds at A, activity goes up When dATP binds at A, activity inhibited overall
Specificity site (S) controls which substrates can be turned over ATP at A + ATP or dATP at S : pyrimidines only dTTP at S : activates reduction of GDP dGTP at S : activates reduction of ADP

06 May 2008Nucleic Acid Metabolism p.37 of 56
dUDP to dUMP(for making dTMP)
dTMP formed at monophosphate level(from dUMP)
dUMP derived three ways: dUDP + ADP dUMP + ATP dUDP + ATP dUTP + ADP
dUTP + H2O dUMP + PPi
dCMP + H2O dUMP + NH4+

06 May 2008Nucleic Acid Metabolism p.38 of 56
Thymidylate synthase reaction (fig.18.15)
dUMP + 5,10-methyleneTHF dTMP + 7,8-dihydrofolate
Unusual THF reaction in that cofactor gets oxidized as well as giving up a carbon CH2 from 5,10-methylene group extra H from C6
So DHF must be reduced back to THF via DHFR and get its methylene back from SHMT
5,10-methylene THF
dihydrofolate

06 May 2008Nucleic Acid Metabolism p.39 of 56
Thymidylate synthase Generally the controlling step in
DNA synthesis because [dTTP] < other [deoxynucleoside triphosphates]
Therefore a target for cancer chemotherapy and other therapies that target rapidly-dividing cells
Enzyme is a 2-layer sandwich
PDB 2G8O58 kDa dimer
E.coli(with dUMP
and cofactor analog)

06 May 2008Nucleic Acid Metabolism p.40 of 56
Thymidylate synthase and drug design
Both folate analogs and dUMP analogs can interfere with (DHFR SHMT dTMP synthase … ) cycle
5-fluorouracil is specific to thymidylate synthase

06 May 2008Nucleic Acid Metabolism p.41 of 56
DHFR Converts DHF to THF:
DHF + NADPH + H+ <->THF + NADP+
SHMT then converts THF to 5,10-methyleneTHF
3-layer sandwich Often the target for drug design
as well Eukaryotic DHFR also catalyzes
folate DHF Prokaryotic DHFR doesn’t;
DHF derived by another mechanism in bacteria
PDB 1KMV20 kDa monomerhuman
folate

06 May 2008Nucleic Acid Metabolism p.42 of 56
Special case:protozoan
TSynth/DHFR Bifunctional enzyme:
Thymidylate synthase Dihydrofolate reductase
Presumably some entropic advantage
Maybe electrostatics too, allowing the negative charges on DHF to tunnel through;but cf. Atreya et al (2003) J.Biol.Chem. 278:28901.
DHFR-TSPDB 1J3K
104 kDa dimer
Plasmodium falciparum

06 May 2008Nucleic Acid Metabolism p.43 of 56
Recovery pathway to dTMP
Deoxythymidine can be phosphorylated by thymidine kinase:deoxythymidine + ATP dTMP + ADP
Labeled thymidine is convenient for monitoring intracellular synthesis of DNA because thymidine enters cells easily
PDB 1E2K73 kDa monomerHerpes simplex virus

06 May 2008Nucleic Acid Metabolism p.44 of 56
Fates of polynucleotides
Nucleic acids hydrolyzed to mononucleotides via nucleases
Mononucleotides are dephosphorylated via nucleotidases and phosphatases
Resulting nucleosides are deglycosylated via nucleosideases or nucleoside phosphorylases
Resulting bases are sent either into salvage pathways or get degraded and excreted

06 May 2008Nucleic Acid Metabolism p.45 of 56
Salvage pathways We can describe them, and we will: but
why do they matter so much? They provide energy savings relative to de novo
synthesis (think of all the ATP we used in making IMP!)
Considerable medical significance to interference with these pathways
Intracellular nucleic acid bases are usually recycled; dietary bases are usually broken down and excess nitrogen excreted

06 May 2008Nucleic Acid Metabolism p.46 of 56
Orotate phosphoribosyl transferase
Principal salvage enzyme for pyrimidines
Orotate + PRPP -> OMP + PPi
OMP can then reenter UMP synthetic pathway (decarboxylation to UMP, then form UDP and CDP)
Same enzyme can aact on other pyrimidines to make nucleotides:Pyr + PRPP -> PyrMP + PPi
PDB 2 PS150 kDa dimerYeast

06 May 2008Nucleic Acid Metabolism p.47 of 56
Pyrimidine interconversions (fig. 18.19) All phosphorylations & dephosphorylations can
and do happen UTP can be aminated to CTP CDP and UDP can be reduced to dCDP and
dUDP dCMP can deaminate to dUMP Cytidine can be converted to uridine dUMP can be methylated to dTMP

06 May 2008Nucleic Acid Metabolism p.48 of 56
Purine nucleotide salvage
Two phosphoribosyl transferases convert adenine, guanine, and hypoxanthine to AMP, GMP, and IMP
Adenine phosphoribosyl transferase is specific
HGPRT accepts both hypoxanthine and guanine
Hypoxanthine-guaninephosphoribosyltransferasePDB 1FSG102 kDa tetramerdimer shownToxoplasma gondii

06 May 2008Nucleic Acid Metabolism p.49 of 56
Purine Interconnections(fig. 18.18)
All phosphorylations and dephosphorylations can and do occur
ADP and GDP can be reduced to dADP and dGDP
AMP can deaminated to IMP (new) IMP can be aminated to AMP IMP can oxidized to XMP XMP can be aminated to GMP Guanine, adenine can be
phosphoribosylated to GMP and AMP

06 May 2008Nucleic Acid Metabolism p.50 of 56
Fates of CMP and cytidine
CMP’s phosphate can be hydrolyzed off
That’s followed by deamination of cytidine to make uridine Catalyzed by cytidine
deaminase Another sandwich
protein
Cytidine deaminasePDB 2FR564 kDa tetramerMouse

06 May 2008Nucleic Acid Metabolism p.51 of 56
Hydrolysis of U, dU and dT
Glycosidic bond in uridine or thymidine is hydrolyzed by phosphate: Uridine + Pi -> -D-ribose-1-P +
uracil Enyzme is uridine
phosphorylase Similar enzyme handles
deoxyuridine Similar reaction using
thymidine phosphorylase yields thymine + -D-deoxyribose-1-P
Uridine phosphorylasePDB 1RXY167 kDa hexamerDimer shownE.coli

06 May 2008Nucleic Acid Metabolism p.52 of 56
Uracil to acetyl CoA;thymine to succinyl CoA Reduced to dihydrouracil and
dihydrothymine Hydrated and ring-opened to
ureidopropionate or ureidoisobutyrate
Eliminate bicarbonate and ammonium to yield -alanine or -aminoisobutyrate
Several reactions from there to acetyl CoA and succinyl CoA
Dihydro-pyrimidinasePDB 1GKP302 kDa hexamerThermus

06 May 2008Nucleic Acid Metabolism p.53 of 56
Purine catabolism
Nucleoside or deoxynucleoside + phosphate base + (D)-ribose 1-P
Hypoxanthine and guanine both lead to uric acid as a product
Uric acid is the final excreted nitrogenous compound in primates and birds and some reptiles
Other organisms catabolize it further
Uric acid

06 May 2008Nucleic Acid Metabolism p.54 of 56
Uric acid to allantoin
Urate oxidase:urate + 2H2O + O2 allantoin + H2O2 + CO2
That’s the final product in a lot of mammals, turtles, some insects, gastropods
Other organisms catabolize allantoin further; we’ll talk about that on Thursday
Uric acid
Allantoin
Urate oxidase134 kDa tetramermonomer shown
Aspergillus flavus

06 May 2008Nucleic Acid Metabolism p.55 of 56
Lesch-Nyhan syndrome
Complete lack of hypoxanthine-guanine phosphoribosyl transferase
So hypoxanthine and guanine are degraded to uric acid rather than being built back up into IMP and GMP
Leads to dangerous buildup of uric acid in nervous tissue
Neurological effects are severe and poorly understood
Michael Lesch
William Nyhan

06 May 2008Nucleic Acid Metabolism p.56 of 56
Gout
Accumulation of sodium urate and uric acid, both of which are only moderately soluble
Arises from inadequate (~10%) functionality of HGPRT, so that urate accumulates in peripheral tissues, particularly the feet
Sodium urate crystalsaccumulating
Sodium urate
Benjamin Franklin(celebrated gout sufferer)