Y chromosome haplogroups and prostate cancer in populations … · 2017. 8. 26. · Hum Genet...
Transcript of Y chromosome haplogroups and prostate cancer in populations … · 2017. 8. 26. · Hum Genet...
-
Hum Genet (2012) 131:1173–1185
DOI 10.1007/s00439-012-1139-5
ORIGINAL INVESTIGATION
Y chromosome haplogroups and prostate cancer in populations of European and Ashkenazi Jewish ancestry
Zhaoming Wang · Hemang Parikh · Jinping Jia · Timothy Myers · Meredith Yeager · Kevin B. Jacobs · Amy Hutchinson · Laurie Burdett · Arpita Ghosh · Michael J. Thun · Susan M. Gapstur · W. Ryan Diver · Jarmo Virtamo · Demetrius Albanes · Geraldine Cancel-Tassin · Antoine Valeri · Olivier Cussenot · Kenneth OYt · Ed Giovannucci · Jing Ma · Meir J. Stampfer · J. Michael Gaziano · David J. Hunter · Ana Dutra-Clarke · Tomas KirchhoV · Michael Alavanja · Laura B. Freeman · Stella Koutros · Robert Hoover · Sonja I. Berndt · Richard B. Hayes · Ilir Agalliu · Robert D. Burk · Sholom Wacholder · Gilles Thomas · Laufey Amundadottir
Received: 22 November 2011 / Accepted: 4 January 2012 / Published online: 24 January 2012© The Author(s) 2012. This article is published with open access at Springerlink.com
Abstract Genetic variation on the Y chromosome has notbeen convincingly implicated in prostate cancer risk. Tocomprehensively analyze the role of inherited Y chromo-some variation in prostate cancer risk in individuals of Euro-pean ancestry, we genotyped 34 binary Y chromosomemarkers in 3,995 prostate cancer cases and 3,815 controlsubjects drawn from four studies. In this set, we identiWednominally signiWcant association between a rare haplogroup,
E1b1b1c, and prostate cancer in stage I (P = 0.012,OR = 0.51; 95% conWdence interval 0.30–0.87). Populationsubstructure of E1b1b1c carriers suggested Ashkenazi Jew-ish ancestry, prompting a replication phase in individuals ofboth European and Ashkenazi Jewish ancestry. The associa-tion was not signiWcant for prostate cancer overall in studiesof either Ashkenazi Jewish (1,686 cases and 1,597 controlsubjects) or European (686 cases and 734 control subjects)ancestry (Pmeta = 0.078), but a meta-analysis of stage I and IIstudies revealed a nominally signiWcant association withprostate cancer risk (Pmeta = 0.010, OR = 0.77; 95% conW-dence interval 0.62–0.94). Comparing haplogroup frequen-cies between studies, we noted strong similarities betweenthose conducted in the US and France, in which the majority
Z. Wang and H. Parikh are co-Wrst authors.
Electronic supplementary material The online version of this article (doi:10.1007/s00439-012-1139-5) contains supplementary material, which is available to authorized users.
Z. Wang · H. Parikh · J. Jia · T. Myers · M. Yeager · K. B. Jacobs · A. Hutchinson · L. Burdett · A. Ghosh · D. Albanes · M. Alavanja · L. B. Freeman · S. Koutros · R. Hoover · S. I. Berndt ·S. Wacholder · L. AmundadottirDivision of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA
Z. Wang · T. Myers · M. Yeager · K. B. Jacobs · A. Hutchinson · L. BurdettCore Genotyping Facility, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, USA
H. Parikh · J. Jia · T. Myers · L. AmundadottirLaboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20877, USA
M. J. Thun · S. M. Gapstur · W. Ryan DiverEpidemiology Research Program, American Cancer Society, Atlanta, GA 30303, USA
J. VirtamoDepartment of Chronic Disease Prevention, National Institute for Health and Welfare, 00300 Helsinki, Finland
G. Cancel-Tassin · A. Valeri · O. CussenotCentre de Recherche pour les Pathologies Prostatiques (CeRePP), Hôpital Tenon, Assistance Publique-Hôpitaux de Paris, 75020 Paris, France
K. OYt · A. Dutra-Clarke · T. KirchhoVClinical Genetics Service, Department of Medicine, Memorial Sloan-Kettering Cancer Center, Box 192, 1275 York Avenue, New York, NY 10065, USA
E. Giovannucci · J. Ma · M. J. Stampfer · J. Michael GazianoChanning Laboratory, Division of Preventive Medicine, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA
D. J. HunterProgram in Molecular and Genetic Epidemiology, Department of Epidemiology, Harvard School of Public Health, Boston, MA 02115, USA
T. KirchhoV · R. B. HayesDivision of Epidemiology, Department of Environmental Medicine, New York University School of Medicine, New York, NY 10016, USA
123
http://dx.doi.org/10.1007/s00439-012-1139-5
-
1174 Hum Genet (2012) 131:1173–1185
of men carried R1 haplogroups, resembling NorthwesternEuropean populations. On the other hand, Finns had aremarkably diVerent haplogroup distribution with a prepon-derance of N1c and I1 haplogroups. In summary, our resultssuggest that inherited Y chromosome variation plays a lim-ited role in prostate cancer etiology in European populationsbut warrant follow-up in additional large and well character-ized studies of multiple ethnic backgrounds.
Introduction
Family and twin studies have shown that prostate cancerhas a clear heritable component which may be among thehighest of all cancer types (Amundadottir et al. 2004; Lich-tenstein et al. 2000), Over the last few years, genome wideassociation studies (GWAS) have successfully identiWedgermline variants conferring risks of prostate cancer at over45 loci (Amundadottir et al. 2006; Chung and Chanock2011; Eeles et al. 2008, 2009; Gudmundsson et al. 2007a,b, 2008, 2009; Haiman et al. 2007; Kote-Jarai et al. 2011;Schumacher et al. 2011; Takata et al. 2010; Thomas et al.2008; Yeager et al. 2007, 2009). These studies have notimplicated variants on the Y chromosome in the risk ofprostate cancer, possibly due to the fact that very few Ychromosome SNPs have been included on most genotypingchips used to date. Several groups have speciWcally investi-gated the role of Y chromosome haplogroups in prostatecancer risk. Many of these studies are inconclusive due tothe small number of samples and/or markers used. One of
the larger studies was conducted within the multi-ethniccohort (MEC) using samples from prostate cancer casesand control subjects drawn from four ethnic groups. Of the41 haplogroups observed, one was signiWcantly associatedwith prostate cancer in Japanese men (Paracchini et al.2003) but this association was not replicated in a separatestudy from Korea (Kim et al. 2007). No association wasseen between Y haplogroups and prostate cancer in a largeSwedish study (Lindstrom et al. 2008).
The Y chromosome contains the largest non-recombiningregion in the human genome, spanning almost the entirelength of the chromosome. This region is called the non-recombining Y (NRY) or the male-speciWc Y (MSY)(Rozen et al. 2003). In the absence of recombination, theNRY passes mostly unchanged from father to son andobserved mutations reXect the evolutionary history of the Ychromosome. Binary markers can be used to classify Ychromosomes into haplogroups organized by a phylogenetictree. A Wrst generation phylogeny of the tree was publishedin 2002 by the Y Chromosome Consortium (2002) and fur-ther revised in 2008 (Karafet et al. 2008). The Y chromo-some tree now consists of over 300 haplogroups organizedinto 20 major groups or clades (Karafet et al. 2008).
Multiple lines of evidence support a possible role forgenes on the Y chromosome in prostate cancer etiology.Loss of the Y chromosome is one of the most frequent cyto-genetic change seen in prostate tumors and may be an earlyevent in tumorigenesis (Brothman et al. 1999; Jordan et al.2001). In support of the previous assertion, chromosometransfer studies indicate that the human Y chromosome sup-presses tumorigenicity of human prostate cell lines in vivoimplying that it may harbor gene(s) with tumor suppressorfunction (Vijayakumar et al. 2005). Based on the essentialrole of the Y chromosome in secondary sexual diVerentia-tion and its potential role in disease pathogenesis, particu-larly related to the secondary sex organs, we explored thisgenomic region to investigate whether germline variation onthis chromosome plays a role in prostate cancer risk.
Results
We analyzed 7,810 men from the Cancer Genetic Markersof Susceptibility (CGEMS) scan in stage I of this study. Ofthe 34 chromosome Y markers genotyped, 26 wereobserved in our sample (8 markers were monomorphic).With such a sample size, we were able to accurately charac-terize and estimate the Y chromosome frequency distribu-tion in populations of European ancestry for 28haplogroups including three combined groups (R1b1b +R1b*, R1a + R1* and I2b + I2c) as the leaf nodes of theNRY tree (Fig. 1a). Stage I had 41, 76 and 95% power todetect an association with an odds ratio of 1.3 and a MAF
I. Agalliu · R. D. BurkDepartment of Epidemiology and Population Health, Albert Einstein College of Medicine, Bronx, NewYork, NY 10461, USA
R. D. BurkDepartment of Pediatrics, Albert Einstein College of Medicine, Bronx, NewYork, NY 10461, USA
R. D. BurkDepartment of Microbiology & Immunology, Albert Einstein College of Medicine, Bronx, NewYork, NY 10461, USA
R. D. BurkDepartment of Obstetrics, Gynecology and Women’s Health, Albert Einstein College of Medicine, Bronx, NewYork, NY 10461, USA
G. ThomasSynergie-Lyon-Cancer, Universite Lyon 1, Centre Leon Berard, 69373 Lyon Cedex 08, France
L. Amundadottir (&)Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Gaithersburg, MD 20877, USAe-mail: [email protected]
123
-
Hum Genet (2012) 131:1173–1185 1175
Fig. 1 Chromosome Y haplogroup tree and frequency distribution incontrol subjects of European ancestry in Stage I. a Chromosome Y treeshowing genotyped markers in black and those not genotyped in lightgrey. Haplogroup names are according to the International Society ofGenetic Genealogy (ISOGG) 2011 update. The arrow points to themutational event which gave rise to the E1b1b1c haplogroup. Stage Istudies are the following: CPS-II American Cancer Society Cancer
Prevention Study II, ATBC Alpha-Tocopherol, Beta-Carotene CancerPrevention Study, CeRePP Centre de Recherche pour les PathologiesProstatiques, and PLCO Prostate, Lung Colorectal and Ovarian CancerScreening Trial. b The circle plots show frequencies for haplogroupswith a derived frequency of 5% or higher in diVerent colors for eachStage I cohort (remaining haplogroups are combined in one groupshown in black)
M42
M60
P97
M168
P143
M203
M174
M96
M132
M180
M78
M81
M123
M130
M377
M285
M201 P15
M522
M26
M307
M172
M170
M304
M9
M70
M20
M526M242
M74
M214
M46
M207
M173 M18
M269
Haplogroup CPS-II ATBC CeRePP PLCO
Q 0.005 0.001 0.004 0.003
R1b1a2 0.462 0.048 0.345 0.488 R1b1b+R1b* 0.083 0 0.297 0.025
R1b1c1 0 0 0 0
R1a+R1* 0.090 0.060 0.024 0.127
R2 0 0 0.002 0.002
O 0 0.032 0 0.001
N*+N1a+N1b 0.001 0.005 0 0
N1c 0.008 0.556 0.004 0.017
L 0 0 0.002 0
T1 0.003 0 0.010 0
J2 0.038 0.001 0.059 0.038
J1 0.029 0 0.002 0.010
I1/O1a1 0.112 0.276 0.081 0.139
I2b+I2c 0.061 0.012 0.049 0.065
I2a1a 0.004 0.001 0.010 0
G2c 0.003 0 0 0.001
G1 0.001 0.001 0 0.001
G2a 0.030 0.005 0.041 0.032
E1b1b1b1 0 0 0.008 0.005
E1b1b1a1 0.028 0.002 0.042 0.024
E1b1b1c 0.020 0 0.010 0.007
C 0.003 0 0 0.001
B 0 0 0 0
D 0 0 0 0
E1a 0.001 0 0 0
E1b1a1a1 0.001 0 0 0
CPS-II ATBC PLCOCeRePP
E1b1b1a1 G2a I Others
A 0 0 0 0.001
A
B
M267
M343
R1N1cJ2
M231
M215
M89
P177
M128+P43
M175
M513
M479
M429
M335
L416/L596
123
-
1176 Hum Genet (2012) 131:1173–1185
of 0.02, 0.05 and 0.10, respectively (assuming prostate can-cer prevalence of 1.5067% and alpha of 0.05) (http://seer.cancer.gov/csr/1975_2007/).
Stage I association analysis
After genotyping quality control based on completion ratesand concordance analysis, a total of 3,995 prostate cancercases and 3,815 control subjects from four studies wereused in the analysis (1994; Calle et al. 2002; Gohagan et al.2000; Valeri et al. 2003). This included 1,531 men diag-nosed with non-aggressive prostate cancer (Gleason score
-
Hum Genet (2012) 131:1173–1185 1177
Tab
le1
Ass
ocia
tion
of c
hrom
osom
e Y
var
iant
s w
ith r
isk
of p
rost
ate
canc
er (
Stag
e I)
Res
ults
fro
m th
e un
cond
itio
nal l
ogis
tic r
egre
ssio
n of
the
geno
type
s ge
nera
ted
in a
tota
l of
3,99
5 in
divi
dual
s w
ith p
rost
ate
canc
er a
nd 3
815
cont
rol s
ubje
cts
are
show
n. T
he a
naly
sis
was
adj
uste
d fo
r ag
e in
10-
year
cat
e-go
ries
, stu
dy a
nd o
ne p
rinc
ipal
com
pone
nt o
f po
pula
tion
str
atiW
cati
on
na*
no r
s nu
mbe
r, O
R o
dds
rati
o, C
I 95
% c
onW
denc
e in
terv
ala
Hap
logr
oup
bein
g te
sted
bas
ed o
n th
e Y
Chr
omos
ome
Con
sort
ium
and
ISO
GG
201
1 no
men
clat
ure
bC
hrY
locu
s/m
arke
r na
me
cN
CB
I db
SNP
iden
tiW
erd
NC
BI
hum
an g
enom
e bu
ild
36 lo
catio
ne
Anc
estr
al a
llel
e, d
eriv
ed a
llel
ef
Con
trol
s, c
ases
gM
inor
all
ele
freq
uenc
y in
con
trol
and
cas
e pa
rtic
ipan
tsh
Sco
re te
st (
1df)
iF
or th
e no
nagg
ress
ive
mod
el, s
ubje
cts
in tw
o ca
tego
ries
(“F
PC
C”
and
“age
<50
”) c
ould
not
be
used
bec
ause
we
ther
e ar
e no
cas
e su
bjec
ts in
eit
her
cate
gory
. The
refo
re, t
he to
tal n
umbe
r of
con
trol
s is
sm
alle
r th
an in
the
over
all a
nd a
ggre
ssiv
e m
odel
s
Hap
logr
oupa
Loc
usb
rs n
umbe
rcL
ocat
iond
Alle
lese
All
case
sN
onag
gres
sive
cas
esA
ggre
ssiv
e ca
ses
Sub
ject
sfM
AFg
Ph
OR
Sub
ject
sf,i
MA
Fg
Ph
OR
Sub
ject
sfM
AF
gP
hO
R
EM
96rs
9306
841
2023
8386
C|G
3739
|390
30.
042|
0.04
30.
638
1.06
(0.
84–1
.32)
3233
|147
60.
039|
0.02
90.
188
0.78
(0.
54–1
.13)
3739
|211
10.
042|
0.05
60.
242
1.16
(0.
90–1
.50)
E1b
1bM
215
rs20
3265
413
9772
18T
|C38
07|3
983
0.04
2|0.
042
0.74
61.
04 (
0.83
–1.3
0)32
98|1
527
0.03
9|0.
029
0.19
60.
79 (
0.55
–1.1
3)38
07|2
134
0.04
2|0.
055
0.30
31.
14 (
0.89
–1.4
8)
E1b
1b1a
1M
78na
*20
3526
91G
|A37
53|3
948
0.02
3|0.
029
0.11
21.
26 (
0.95
–1.6
7)32
47|1
512
0.02
1|0.
022
0.72
51.
08 (
0.70
–1.6
7)37
53|2
116
0.02
3|0.
036
0.08
21.
33 (
0.96
–1.8
3)
E1b
1b1b
1M
81rs
2032
640
2035
1960
G|A
3243
|337
20.
002|
0.00
10.
079
0.27
(0.
06–1
.29)
2738
|122
30.
001|
0.00
0N
AN
A32
43|1
945
0.00
2|0.
001
0.19
20.
36 (
0.07
–1.7
7)
E1b
1b1c
M12
3na
*20
2239
74C
|T38
14|3
992
0.01
1|0.
005
0.01
20.
51 (
0.30
–0.8
7)33
06|1
531
0.01
1|0.
003
0.01
70.
33 (
0.13
–0.8
6)38
14|2
140
0.01
1|0.
007
0.09
10.
59 (
0.32
–1.0
9)
FM
89rs
2032
652
2037
6701
A|G
3813
|399
40.
043|
0.04
40.
641
1.05
(0.
85–1
.31)
3304
|153
10.
040|
0.03
10.
267
0.82
(0.
58–1
.16)
3813
|214
10.
043|
0.05
70.
255
1.16
(0.
90–1
.49)
GM
201
rs20
3263
613
5369
23G
|T38
04|3
987
0.02
9|0.
026
0.52
80.
92 (
0.70
–1.2
0)32
96|1
529
0.02
7|0.
026
0.99
51.
00 (
0.68
–1.4
8)38
04|2
136
0.02
9|0.
029
0.38
80.
87 (
0.62
–1.2
0)
G1
M28
5rs
1344
7378
2115
1128
G|C
3807
|399
00.
001|
0.00
10.
973
1.02
(0.
26–4
.11)
3300
|152
80.
001|
0.00
20.
440
1.86
(0.
38–9
.12)
3807
|214
00.
001|
0.00
0N
AN
A
G2a
P15
na*
2165
3414
G|A
3791
|394
80.
026|
0.02
30.
328
0.87
(0.
65–1
.16)
3302
|152
90.
024|
0.02
20.
714
0.92
(0.
61–1
.41)
3791
|209
80.
026|
0.02
50.
310
0.84
(0.
59–1
.18)
G2c
M37
7na
*13
5368
27A
|G37
50|3
950
0.00
2|0.
002
0.41
01.
54 (
0.55
–4.3
5)32
41|1
508
0.00
2|0.
002
0.84
31.
16 (
0.28
–4.8
5)37
50|2
120
0.00
2|0.
002
0.47
41.
56 (
0.46
–5.2
9)
IM
170
rs20
3259
713
3571
86A
|C38
11|3
990
0.20
3|0.
221
0.05
91.
11 (
1.00
–1.2
4)33
02|1
529
0.21
3|0.
247
0.04
21.
17 (
1.01
–1.3
5)38
11|2
139
0.20
3|0.
204
0.05
81.
14 (
1.00
–1.3
1)
I1/O
1a1
M30
7rs
1344
7354
2116
0339
G|A
3804
|398
80.
150|
0.15
40.
648
1.03
(0.
91–1
.17)
3298
|152
50.
161|
0.17
60.
737
1.03
(0.
87–1
.22)
3804
|214
10.
150|
0.13
60.
249
1.10
(0.
94–1
.29)
I2a1
aM
26rs
2032
629
2032
5209
C|T
3719
|387
50.
003|
0.00
20.
298
0.62
(0.
25–1
.53)
3215
|145
30.
002|
0.00
30.
537
1.51
(0.
41–5
.58)
3719
|210
70.
003|
0.00
20.
117
0.41
(0.
13–1
.30)
JM
304
rs13
4473
5221
1592
41A
|C37
42|3
922
0.04
8|0.
048
0.63
51.
05 (
0.85
–1.3
1)32
34|1
492
0.04
6|0.
035
0.30
20.
84 (
0.60
–1.1
7)37
42|2
116
0.04
8|0.
059
0.37
91.
12 (
0.87
–1.4
3)
J2M
172
rs20
3260
413
4790
28T
|G37
80|3
958
0.03
3|0.
033
0.65
71.
06 (
0.82
–1.3
7)32
72|1
516
0.02
8|0.
022
0.69
70.
92 (
0.61
–1.3
9)37
80|2
124
0.03
3|0.
043
0.49
31.
11 (
0.83
–1.4
7)
KM
9rs
3900
2018
9645
C|G
3799
|397
50.
322|
0.34
00.
055
1.10
(1.
00–1
.21)
3294
|152
50.
324|
0.33
80.
299
1.07
(0.
94–1
.22)
3799
|212
90.
322|
0.34
80.
021
1.15
(1.
02–1
.29)
T1
M70
rs20
3267
220
3532
69T
|G37
08|3
904
0.00
3|0.
003
0.92
21.
04 (
0.44
–2.4
7)32
02|1
492
0.00
2|0.
001
0.31
52.
92 (
0.33
–25.
65)
3708
|210
10.
003|
0.00
40.
882
0.93
(0.
37–2
.32)
NM
231
rs93
4127
813
9791
18C
|T37
69|3
946
0.13
9|0.
136
0.66
40.
96 (
0.80
–1.1
5)32
62|1
505
0.16
0|0.
189
0.56
50.
94 (
0.76
–1.1
7)37
69|2
122
0.13
9|0.
062
0.07
60.
78 (
0.60
–1.0
3)
N1c
M46
/Tat
rs34
4421
2613
4319
77T
|C38
09|3
992
0.13
7|0.
134
0.60
40.
95 (
0.80
–1.1
4)33
02|1
530
0.15
7|0.
185
0.51
40.
93 (
0.75
–1.1
6)38
09|2
140
0.13
7|0.
062
0.05
80.
77 (
0.59
–1.0
1)
NO
M21
4rs
2032
674
1398
1319
A|G
3805
|397
70.
139|
0.13
50.
676
0.96
(0.
80–1
.15)
3297
|152
30.
159|
0.18
60.
578
0.94
(0.
76–1
.17)
3805
|213
50.
139|
0.06
20.
058
0.77
(0.
59–1
.01)
PM
74rs
2032
635
2034
9155
C|T
3547
|376
60.
484|
0.49
40.
272
1.06
(0.
96–1
.18)
3047
|141
80.
491|
0.45
50.
959
1.00
(0.
86–1
.16)
3547
|204
10.
484|
0.42
70.
282
1.07
(0.
95–1
.21)
QM
242
rs81
7902
113
5279
76C
|T37
19|3
856
0.00
4|0.
004
0.88
61.
05 (
0.51
–2.1
9)32
29|1
487
0.00
4|0.
003
0.84
20.
89 (
0.27
–2.8
8)37
19|2
058
0.00
4|0.
005
0.35
31.
47 (
0.65
–3.3
5)
RM
207
rs20
3265
814
0913
77T
|C37
45|3
908
0.48
6|0.
499
0.08
81.
09 (
0.99
–1.2
1)32
38|1
497
0.49
0|0.
453
0.66
30.
97 (
0.84
–1.1
2)37
45|2
098
0.48
6|0.
436
0.09
41.
11 (
0.98
–1.2
5)
R1
M17
3rs
2032
624
1353
5818
A|C
3691
|387
60.
477|
0.48
70.
144
1.08
(0.
97–1
.19)
3189
|148
10.
500|
0.46
70.
783
0.98
(0.
85–1
.13)
3691
|207
60.
477|
0.42
30.
160
1.09
(0.
97–1
.23)
R1b
M34
3rs
9786
184
2947
824
C|A
3805
|397
60.
441|
0.42
80.
114
0.92
(0.
84–1
.02)
3297
|152
50.
410|
0.37
10.
673
0.97
(0.
84–1
.12)
3805
|213
10.
441|
0.49
50.
193
0.92
(0.
82–1
.04)
R1b
1a2
M26
9rs
9786
153
2114
8755
T|C
3489
|363
40.
437|
0.42
10.
054
0.90
(0.
81–1
.00)
3029
|138
60.
405|
0.36
50.
719
0.97
(0.
84–1
.13)
3489
|195
90.
437|
0.49
10.
093
0.90
(0.
79–1
.02)
123
-
1178 Hum Genet (2012) 131:1173–1185
with risk of aggressive prostate cancer (Pmeta = 0.28). InStage II, we had 60% power to detect a variant with 13%MAF and an OR of 0.8 in the Ashkenazi Jewish sample set,but only 25% power to detect a variant with 3% MAF andan OR of 0.7 in the European American sample set.
Haplogroup frequency and population distribution
Y chromosome haplogroup frequency distribution in con-trols from each of the four study populations from phase Iwas summarized and compared in Fig. 1b. Two of the stud-ies, namely CPS-II and PLCO, include subjects from conti-nental USA. Their haplogroup frequencies are very similarwith an average diVerence of 0.8% and a maximum diVer-ence of 5.8% for the combined category of haplogroupsR1b1b + R1b*. The CeRePP study, conducted in France, isrelatively similar to the US studies with an average haplo-group frequency diVerence of 2.2%, and a maximum diVer-ence of 24.3% for the combined group of R1b1b + R1b*.The greatest diVerence in frequency was seen for ATBC, aFinnish study, with an average haplogroup frequency diVer-ence of 5.2% and a maximum diVerence of 54.4%. Thisstems from a very high frequency of haplogroup N1c in thisstudy (55.6%), while it is infrequent in the other three stud-ies from the US and France (0.8% in CPS-II, 1.7% inPLCO and 0.4% in CeRePP). Second, R1b, the most fre-quent haplogroup overall, is seen in over 50% of subjects inPLCO, CPS-II and CeRePP but only in 4.8% of Finnishsubjects. The third largest diVerence was noted for haplo-group I1 which was more common in Finns at 27.6%, ascompared to 13.9% in PLCO, 11.2% in CPS-II and only8.1% in CeRePP.
Haplogroup E1b was observed at low frequencies in allstudies and its sub lineage E1b1b1c was seen in approxi-mately 1–2% of subjects from the two US studies (PLCOand CPS-II) and the French study (CeRePP), whereas itwas absent from the Finnish study (ATBC). Other haplo-groups were absent or rare in the four studies.
Discussion
In this study, we explored the role of germline Y chromo-some variation in prostate cancer risk. Previous studieshave not analyzed such a large sample size with as manymarkers in individuals of European ancestry. Because ofthe threshold for MAF chosen for this study (¸1%), we hadlimited capacity to detect risk variants with low to mediumfrequency and eVect sizes. Prostate cancer GWAS to datehave used arrays with limited coverage on the Y chromo-some. As an example, in CGEMS, of the approximately500,000 SNPs genotyped in stage I, only ten Y chromo-some markers passed quality control assessment and wereincluded in the primary analysis; this limited set of variantson the Y chromosome included only four that mark chro-mosome Y haplogroups (Thomas et al. 2008; Yeager et al.2007, 2009). Other published prostate cancer GWAS stud-ies have reported on a similar fraction of Y variants (Amun-dadottir et al. 2006; Chung and Chanock 2011; Eeles et al.
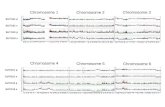
Fig. 2 Population substructure analysis by principal component anal-ysis and comparison to CGEMS prostate cancer GWAS. a shows thedistribution of the Wrst two principal components, EV1 and EV2, forcarriers of E1b1b1c (Wlled squares) and R1b1a2 (open circles) haplo-groups in Stage I. Circles and squares denote eigenvalues from PCAanalysis for each individual. The distribution of EV1 and EV2 for allStage I subjects is shown in b. Studies are designed by diVerent colors.CPS-II Blood blood derived DNA samples were used for genotyping,CPS-II Buccal buccal derived DNA samples were used for genotyping.DNA samples from ATBC, CeRePP and PLCO were all derived fromblood. Individuals of inferred Ashkenazi Jewish ancestry are circled.PCA results were performed by EIGENSTRAT in CGEMS prostatecancer GWAS (Thomas et al. 2008; Yeager et al. 2007, 2009)
E1b1b1c
R1b1a2
-0.01
0
0.01
0.02
0.03
0.04
0.05
0.03
EV1
EV
2
-0.02 -0.01 0 0.01 0.02
-0.01
0
0.01
0.02
0.03
0.04
0.05
0.03
EV1
EV
2
ATBCCPS-II:BloodCPS-II:Buccal
CeRePP
PLCO
AJ alike cluster
-0.02 -0.01 0 0.01 0.02
A
B
123
-
Hum Genet (2012) 131:1173–1185 1179
Tab
le2
Rep
licat
ion
anal
ysis
of
nota
ble
chro
mos
ome
Y s
igna
ls in
pro
stat
e ca
ncer
stu
dies
of
Ash
kena
zi a
nd E
urop
ean
desc
ent (
Stag
e II
)
Res
ults
fro
m th
e un
cond
ition
al lo
gist
ic r
egre
ssio
n of
the
geno
type
s ge
nera
ted
in S
tage
II
are
show
n. T
he a
naly
sis
was
adj
uste
d fo
r ag
e in
10-
year
cat
egor
ies
OR
odd
s ra
tio,
CI
95%
conW
denc
e in
terv
ala
Hap
logr
oup
bein
g te
sted
bas
ed o
n th
e Y
Chr
omos
ome
Con
sort
ium
and
IS
OG
G 2
011
nom
encl
atur
eb
Chr
Y lo
cus/
mar
ker
nam
ec
Con
trol
s, c
ases
dM
inor
all
ele
freq
uenc
y in
con
trol
and
cas
e pa
rtic
ipan
tse
Sco
re te
st (
1df)
for
all
scen
ario
s ex
cept
for
agg
ress
ive
case
s fr
om H
PFS
whi
ch is
bas
ed o
n a
Fish
er e
xact
test
(1d
f)
Stud
yA
ll c
ases
Non
aggr
essi
ve c
ases
Agg
ress
ive
case
s
Sub
ject
scM
AFd
Pe
OR
Sub
ject
scM
AFd
Pe
OR
Sub
ject
scM
AFd
Pe
OR
E1b
1b1c
a (M
123b
)
Ein
stei
n12
14|9
280.
138|
0.11
70.
146
0.82
(0.
63–1
.07)
1214
|413
0.13
8|0.
097
0.02
40.
66 (
0.45
–0.9
5)12
14|4
560.
138|
0.13
40.
693
0.94
(0.
68–1
.29)
MSK
CC
375|
750
0.12
8|0.
123
0.60
80.
87 (
0.52
–1.4
6)37
5|21
20.
128|
0.09
00.
956
0.96
(0.
47–1
.94)
375|
362
0.12
8|0.
146
0.95
40.
98 (
0.53
–1.8
3)
PHS
489|
471
0.02
9|0.
021
0.45
50.
73 (
0.32
–1.6
7)48
9|19
40.
029|
0.01
50.
379
0.57
(0.
16–2
.02)
489|
167
0.02
9|0.
024
0.78
00.
85 (
0.28
–2.6
3)
HPF
S24
4|21
50.
033|
0.02
30.
540
0.70
(0.
23–2
.18)
244|
138
0.03
3|0.
036
0.84
81.
12 (
0.36
–3.4
9)24
4|44
0.03
3|0.
000
0.21
00.
00 (
0.00
–1.7
7)
R1b
1a2
(M26
9b)
Ein
stei
n12
10|9
120.
095|
0.11
50.
121
1.25
(0.
94–1
.67)
1210
|408
0.09
5|0.
120
0.14
71.
30 (
0.91
–1.8
7)12
10|4
460.
095|
0.10
80.
390
1.17
(0.
82–1
.69)
MSK
CC
373|
739
0.11
0|0.
118
0.93
61.
02 (
0.61
–1.7
137
3|20
60.
110|
0.12
10.
981
0.99
(0.
51–1
.93)
373|
358
0.11
0|0.
126
0.84
11.
06(0
.59–
1.92
)
PHS
482|
463
0.50
0|0.
531
0.32
11.
14 (
0.88
–1.4
7)48
2|19
00.
500|
0.55
30.
238
1.23
(0.
87–1
.72)
482|
164
0.50
0|0.
537
0.43
01.
15 (
0.81
–1.6
5)
AH
S11
59|5
710.
589|
0.57
80.
662
0.96
(0.
78–1
17)
1159
|326
0.58
9|0.
558
0.32
00.
88(0
.66–
1.09
)11
59|8
00.
589|
0.61
20.
703
1.09
(0.
69–1
.74)
123
-
1180 Hum Genet (2012) 131:1173–1185
2008, 2009; Gudmundsson et al. 2007a, b, 2008, 2009;Haiman et al. 2007; Kote-Jarai et al. 2011; Schumacheret al. 2011; Takata et al. 2010; Thomas et al. 2008; Yeageret al. 2007, 2009).
One haplogroup of interest was noted in phase I of ourstudy; the E1b1b1c haplogroup was nominally signiWcantin the overall prostate cancer and non-aggressive prostatecancer groups. The marker that denotes this haplogroup islocated in the last intron of the taxilin gamma 2 pseudogene(TXLNG2P) on chromosome Yq11.222. This haplogroupwas analyzed in a second phase using replication studies ofEuropean and Ashkenazi Jewish ancestry along with amore common haplogroup, R1b1a2. Neither haplogroupwas signiWcantly associated with overall prostate cancerrisk in stage II. A meta-analysis of stage I and stage IIresults yielded a P value of 0.010 for the E1b1b1c haplo-group. Although nominally signiWcant, this P value is unre-markable in comparison with the rigorous thresholdrequired for signiWcance in GWAS studies (WellcomeTrust Case Control Consortium 2007), suggesting that fur-ther studies are required to establish this association.Although our analysis does not provide strong evidence fora relationship between variation in the Y chromosome andprostate cancer, it can be argued that the appropriate statis-tical threshold to be applied to a study of approximately 30markers should not be as stringent as a GWAS threshold.However, the probability of false-positive Wndings is high,even in a study of our size and power (Wacholder et al.2004) especially in the Wrst stage where E1b1b1c haplo-group frequency was very low. In addition, we cannotexclude a chance Wnding due to population stratiWcation.
Our study represents the largest analysis to date of a pos-sible association between Y chromosome variants and pros-tate cancer. The role of germline variation on the Ychromosome had been assessed previously, but with limitedsample and/or marker sets. One of the most complete stud-ies published was conducted within the MEC (Paracchiniet al. 2003). Four ethnic groups with a total of 930 casesand 1,208 control subjects were included. One of the 41haplogroups observed in the study was signiWcantly associ-ated with prostate cancer risk in Japanese men with aP value of 0.02 (Paracchini et al. 2003). Despite the largeoverall sample set in this study, each ethnic group only con-sisted of approximately 100–150 case–control pairs, limit-ing power considerably. No haplogroups were signiWcantlyassociated with prostate cancer risk in a small Korean studythat assessed 14 markers in approximately 106 cases and110 control subjects, including the haplogroup reported inthe MEC study (Kim et al. 2007). Lack of an associationbetween Y haplogroups and prostate cancer was alsoreported in a Swedish study assessing Wve ChrY markers in1,452 cases and 779 control subjects of N-European back-ground (Lindstrom et al. 2008). Our results appear to con-
Wrm an overall lack of importance for germline variants onthe Y chromosome and prostate cancer risk.
Frequencies of Y chromosome haplogroups vary consid-erably between diVerent geographical regions and ethnicgroups, and have turned out to be informative in studies ofhuman evolution and migration. In Europe, marked diVer-ences in haplogroup frequencies are observed betweencountries in Northeastern, Northwestern, Southwestern,Southeast and Central Europe (Wiik 2008). In addition, theAshkenazi Jewish community has a speciWc pattern that isreminiscent of non-Ashkenazi Jewish communities in theNear East (Behar et al. 2004). We observed a diVerent distri-bution of major haplogroups in subjects of NorthwesternEuropean ancestry (represented by the majority of subjectsfrom the US in PLCO and CPS-II), Northeastern Europeanancestry (represented by Finnish subjects in ATBC) andWestern/Central European ancestry (represented by Frenchsubjects in CeRePP). Haplogroups in the US and Frenchstudies can mostly be accounted for by the R and I haplo-group clans with a combined frequency of 81–85%; R1b1a2and I1 were the most common sub branches. The R1 haplo-group clan originated in Eurasia and migrated into Europewhere it divided into two subgroups, R1a (common in East-ern Europe) and R1b (common in Western Europe) (Wiik2008). R1b1a2 shows an East to West gradient in Europeand is very common in Spain, France, UK and Ireland (Bal-aresque et al. 2010). Haplogroup clan I1 appears to haveoriginated in the Balkans and migrated north throughoutEurope (Wiik 2008). It is most common in Scandinavia andNorthwestern Europe and gradually decreases in Central andSouthern Europe (Wiik 2008). Finnish subjects were strik-ingly diVerent from the other three studies with a preponder-ance of N1c (56%) and I1 (28%) haplogroups and few R1bcarriers. The N1c haplogroup is thought to have an Easternor Central Asian origin and probably reached EasternEurope via expansion through Siberia (Rootsi et al. 2007).The frequency of this haplogroup in Finland has beenreported to be 58% (Wiik 2008).
Genotypes in stage II conWrmed the scarcity of E1b1b1cin subjects of European ancestry (1–2%) and revealed ahigher frequency in the two Ashkenazi Jewish studies(13–14%), in line with previous reports (Hammer et al.2009) indicating similar Y chromosome haplogroup fre-quencies in men of Ashkenazi Jewish descent living in theUS and those from Jewish communities in the Middle East.E1b1b1c may have arisen in Northeastern Africa, andmigrated through the Levantine corridor to the Near Eastand Europe (Semino et al. 2004). In a similar manner,haplogroup R1b1a2 was seen in 50–59% of the subjects indiVerent European American studies but only 10–11% inthe two Ashkenazi Jewish studies.
In conclusion, we found limited evidence for an associa-tion between Y chromosome haplogroups and risk of
123
-
Hum Genet (2012) 131:1173–1185 1181
prostate cancer in populations of European and AshkenaziJewish ancestry using a large sample set close to 4,000case–control pairs in Stage I and 2,300 case–control pairsin Stage II. Weak but consistent evidence for a protectiveeVect for haplogroup E1b1b1c was seen in all studies with anominally signiWcant meta-analysis, thus, calling for addi-tional replication eVorts for this haplogroup in populationsof Ashkenazi Jewish and European ancestry. The diVerentfrequencies seen in subjects from the four stage I studiesmay limit power to detect true associations for somebranches of the Y haplogroup tree. Furthermore, correctingfor population substructure based on autosomal SNPs maynot be optimal, as Y chromosome inheritance only reXectsmale lineages that may have somewhat diVerent character-istics throughout human history and population migrationas compared to that of females. Although we cannotexclude a role for all chromosome Y haplogroups in pros-tate cancer etiology, our study has good power to detectcommon alleles with relatively large eVects. Smaller orpopulation speciWc eVects for the haplgroups tested here, orfor other haplogroups, could exist and should be studied bytesting comprehensive sets of chromosome Y haplogroupmarkers in additional studies.
Materials and methods
Study population
Stage I of this study included 3,995 men diagnosed withadenocarcinoma of the prostate and 3,815 control subjectsfrom three case–control studies nested within cohorts andone hospital based case–control study, previously analyzedin stages I and II of the Cancer Genetics Markers of Sus-ceptibility study (CGEMS). Study details have been pub-lished previously (Thomas et al. 2008; Yeager et al. 2007,2009).The cohort studies were: the Prostate, Lung Colorec-tal and Ovarian Cancer Screening Trial (PLCO, subjectsfrom continental USA) (Gohagan et al. 2000); the Ameri-can Cancer Society Cancer Prevention Study II (CPS-II,from continental USA) (Calle et al. 2002) and the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study(ATBC, from Finland) (1994). The case–control study wasthe French Prostate Case–Control Study (CeRePP, Centrede Recherche pour les Pathologies Prostatiques, fromFrance) (Valeri et al. 2003). The number of subjectsincluded from each study is shown in SupplementalTable 1a. We incorporated prostate cancer stage and gradeat diagnosis to distinguish between non-aggressive (Glea-son score
-
1182 Hum Genet (2012) 131:1173–1185
99.75%. Samples were excluded based on a completion rate
-
Hum Genet (2012) 131:1173–1185 1183
Chanock, LTG, DCEG, National Cancer Institute. The content of thispublication does not necessarily reXect the views or policies of theDepartment of Health and Human Services, nor does mention of tradenames, commercial products or organizations imply endorsement bythe US Government.
ConXict of interest All authors report no Wnancial interests orpotential conXicts of interests.
Open Access This article is distributed under the terms of the Crea-tive Commons Attribution License which permits any use, distribution,and reproduction in any medium, provided the original author(s) andsource are credited.
References
Agalliu I, Gern R, Leanza S, Burk RD (2009) Associations of high-grade prostate cancer with BRCA1 and BRCA2 founder muta-tions. Clin Cancer Res 15:1112–1120. doi:10.1158/1078-0432.CCR-08-1822
Alavanja MC, Sandler DP, McMaster SB, Zahm SH, McDonnell CJ,Lynch CF, Pennybacker M, Rothman N, Dosemeci M, Bond AE,Blair A (1996) The Agricultural Health Study. Environ HealthPerspect 104:362–369
Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Krist-jansson K, Arnason S, Gulcher JR, Bjornsson J, Kong A, Thor-steinsdottir U, Stefansson K (2004) Cancer as a complexphenotype: pattern of cancer distribution within and beyond thenuclear family. PLoS Med 1:e65. doi:10.1371/journal.pmed.0010065
Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A,Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB,Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S,Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, BlondalT, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjarts-son D, Gylfason A, Thorleifsson G, Manolescu A, KristjanssonK, Geirsson G, Isaksson H, Douglas J, Johansson JE, Balter K,Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA,Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gul-cher JR, Kong A, Thorsteinsdottir U, Stefansson K (2006) A com-mon variant associated with prostate cancer in European andAfrican populations. Nat Genet 38:652–658. doi:10.1038/ng1808
Balaresque P, Bowden GR, Adams SM, Leung HY, King TE, RosserZH, Goodwin J, Moisan JP, Richard C, Millward A, DemaineAG, Barbujani G, Previdere C, Wilson IJ, Tyler-Smith C, JoblingMA (2010) A predominantly neolithic origin for European pater-nal lineages. PLoS Biol 8:e1000285. doi:10.1371/journal.pbio.1000285
Behar DM, Garrigan D, Kaplan ME, Mobasher Z, Rosengarten D, Ka-rafet TM, Quintana-Murci L, Ostrer H, Skorecki K, Hammer MF(2004) Contrasting patterns of Y chromosome variation in Ashke-nazi Jewish and host non-Jewish European populations. HumGenet 114:354–365. doi:10.1007/s00439-003-1073-7
Brothman AR, Maxwell TM, Cui J, Deubler DA, Zhu XL (1999) Chro-mosomal clues to the development of prostate tumors. Prostate38:303–312. doi:10.1002/(SICI)1097-0045(19990301)38:43.0.CO;2-E
Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCulloughML, Feigelson HS, Thun MJ (2002) The American Cancer Soci-ety Cancer Prevention Study II Nutrition Cohort: rationale, studydesign, and baseline characteristics. Cancer 94:2490–2501.doi:10.1002/cncr.101970
Chen YC, Giovannucci E, Lazarus R, Kraft P, Ketkar S, Hunter DJ(2005) Sequence variants of toll-like receptor 4 and susceptibility
to prostate cancer. Cancer Res 65:11771–11778. doi:10.1158/0008-5472.CAN-05-2078
Chung CC, Chanock SJ (2011) Current status of genome-wide associ-ation studies in cancer. Hum Genet 130:59–78. doi:10.1007/s00439-011-1030-9
Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK,Mulholland S, Leongamornlert DA, Edwards SM, Morrison J,Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC,Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT,Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A,Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, LophatananonA, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC,Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C,Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Eas-ton DF (2008) Multiple newly identiWed loci associated withprostate cancer susceptibility. Nat Genet 40:316–321.doi:10.1038/ng.90
Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G,Muir K, Hopper JL, Henderson BE, Haiman CA, Schleutker J,Hamdy FC, Neal DE, Donovan JL, Stanford JL, Ostrander EA,Ingles SA, John EM, Thibodeau SN, Schaid D, Park JY, SpurdleA, Clements J, Dickinson JL, Maier C, Vogel W, Dork T, Reb-beck TR, Cooney KA, Cannon-Albright L, Chappuis PO, HutterP, Zeegers M, Kaneva R, Zhang HW, Lu YJ, Foulkes WD, En-glish DR, Leongamornlert DA, Tymrakiewicz M, Morrison J,Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, SaundersEJ, Page EC, Sawyer EJ, Edwards SM, Dearnaley DP, HorwichA, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ,Thompson A, Christmas T, Ogden C, Cooper CS, Southey MC,Lophatananon A, Liu JF, Kolonel LN, Le Marchand L, WahlforsT, Tammela TL, Auvinen A, Lewis SJ, Cox A, FitzGerald LM,Koopmeiners JS, Karyadi DM, Kwon EM, Stern MC, Corral R,Joshi AD, Shahabi A, McDonnell SK, Sellers TA, Pow-Sang J,Chambers S, Aitken J, Gardiner RA, Batra J, Kedda MA, Lose F,Polanowski A, Patterson B, Serth J, Meyer A, Luedeke M, SteZ-ova K, Ray AM, Lange EM, Farnham J, Khan H, Slavov C, Mitk-ova A, Cao G et al (2009) IdentiWcation of seven new prostatecancer susceptibility loci through a genome-wide associationstudy. Nat Genet 41:1116–1121. doi:10.1038/ng.450
Gallagher DJ, Vijai J, Cronin AM, Bhatia J, Vickers AJ, Gaudet MM,Fine S, Reuter V, Scher HI, Hallden C, Dutra-Clarke A, Klein RJ,Scardino PT, Eastham JA, Lilja H, KirchhoV T, OYt K (2010)Susceptibility loci associated with prostate cancer progressionand mortality. Clin Cancer Res 16:2819–2832. doi:10.1158/1078-0432.CCR-10-0028
Gohagan JK, Prorok PC, Hayes RB, Kramer BS (2000) The Prostate,Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial ofthe National Cancer Institute: history, organization, and status.Control Clin Trials 21:251S–272S
Gold B, KirchhoV T, Stefanov S, Lautenberger J, Viale A, Garber J,Friedman E, Narod S, Olshen AB, Gregersen P, Kosarin K, OlshA, Bergeron J, Ellis NA, Klein RJ, Clark AG, Norton L, Dean M,Boyd J, OYt K (2008) Genome-wide association study providesevidence for a breast cancer risk locus at 6q22.33. Proc Natl AcadSci USA 105:4340–4345. doi:10.1073/pnas.0800441105
Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjarts-son D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA,Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, XuJ, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M,Saemundsdottir J, Backman VM, Kristjansson K, Tres A, PartinAW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC,Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, CashyJ, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M,Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA,Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR,Thorsteinsdottir U, Kong A, Stefansson K (2007a) Genome-wide
123
http://dx.doi.org/10.1158/1078-0432.CCR-08-1822http://dx.doi.org/10.1158/1078-0432.CCR-08-1822http://dx.doi.org/10.1371/journal.pmed.0010065http://dx.doi.org/10.1371/journal.pmed.0010065http://dx.doi.org/10.1038/ng1808http://dx.doi.org/10.1371/journal.pbio.1000285http://dx.doi.org/10.1371/journal.pbio.1000285http://dx.doi.org/10.1007/s00439-003-1073-7http://dx.doi.org/10.1002/(SICI)1097-0045(19990301)38:4%3c303:AID-PROS6%3e3.0.CO;2-Ehttp://dx.doi.org/10.1002/(SICI)1097-0045(19990301)38:4%3c303:AID-PROS6%3e3.0.CO;2-Ehttp://dx.doi.org/10.1002/cncr.101970http://dx.doi.org/10.1158/0008-5472.CAN-05-2078http://dx.doi.org/10.1158/0008-5472.CAN-05-2078http://dx.doi.org/10.1007/s00439-011-1030-9http://dx.doi.org/10.1007/s00439-011-1030-9http://dx.doi.org/10.1038/ng.90http://dx.doi.org/10.1038/ng.450http://dx.doi.org/10.1158/1078-0432.CCR-10-0028http://dx.doi.org/10.1158/1078-0432.CCR-10-0028http://dx.doi.org/10.1073/pnas.0800441105
-
1184 Hum Genet (2012) 131:1173–1185
association study identiWes a second prostate cancer susceptibilityvariant at 8q24. Nat Genet 39:631–637. doi:ng1999[pii]10.1038/ng1999
Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thor-leifsson G, Manolescu A, Rafnar T, Gudbjartsson D, AgnarssonBA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M,Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, OlafsdottirA, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Mag-nusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, WilenskyRL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y,Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Bor-ch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M,Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK,Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML,Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ,Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Bene-diktsson R, Jonsson E, Einarsson GV, Mayordomo JI, CatalonaWJ, Kiemeney LA, Barkardottir RB, Gulcher JR, ThorsteinsdottirU, Kong A, Stefansson K (2007b) Two variants on chromosome17 confer prostate cancer risk, and the one in TCF2 protectsagainst type 2 diabetes. Nat Genet 39: 977–983. doi:10.1038/ng2062
Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A,Gudbjartsson D, Agnarsson BA, Sigurdsson A, BenediktsdottirKR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, KristinssonKT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S,Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, BreyerJP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stat-tin P, Lindstrom S, Adami HO, McDonnell SK, Schaid DJ, Cunn-ingham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, WileyKE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S,Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, AbenKK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, KristjanssonK, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, SmithJR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kie-meney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U,Kong A, Stefansson K (2008) Common sequence variants on2p15 and Xp11.22 confer susceptibility to prostate cancer. NatGenet 40:281–283. doi:10.1038/ng.89
Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A,Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygs-dottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T,Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B,Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK,van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, FriggeML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Cata-lona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J,Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefans-son K (2009) Genome-wide association and replication studiesidentify four variants associated with prostate cancer susceptibil-ity. Nat Genet 41:1122–1126. doi:10.1038/ng.448
Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Wal-iszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ,Greenway SC, Stram DO, Le Marchand L, Kolonel LN, FrascoM, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, WhittemoreAS, Cooney KA, John EM, Ingles SA, Altshuler D, HendersonBE, Reich D (2007) Multiple regions within 8q24 independentlyaVect risk for prostate cancer. Nat Genet 39:638–644.doi:10.1038/ng2015
Hammer MF, Behar DM, Karafet TM, Mendez FL, Hallmark B, ErezT, Zhivotovsky LA, Rosset S, Skorecki K (2009) Extended Ychromosome haplotypes resolve multiple and unique lineages ofthe Jewish priesthood. Hum Genet 126:707–717. doi:10.1007/s00439-009-0727-5
Jordan JJ, Hanlon AL, Al-Saleem TI, Greenberg RE, Tricoli JV (2001)Loss of the short arm of the Y chromosome in human prostate car-
cinoma. Cancer Genet Cytogenet 124:122–126. pii:S0165-4608(00)00340-X
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL,Hammer MF (2008) New binary polymorphisms reshape and in-crease resolution of the human Y chromosomal haplogroup tree.Genome Res 18:830–838. doi:10.1101/gr.7172008
Kim W, Yoo TK, Kim SJ, Shin DJ, Tyler-Smith C, Jin HJ, Kwak KD,Kim ET, Bae YS (2007) Lack of association between Y-chromo-somal haplogroups and prostate cancer in the Korean population.PLoS One 2:e172. doi:10.1371/journal.pone.0000172
Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, WeischerM, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P,Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE,Hamdy FC, Donovan JL, Pharoah P, Schumacher F, HendersonBE, Stanford JL, Ostrander EA, Sorensen KD, Dork T, AndrioleG, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, ClementsJA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, SchaidD, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R,Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tym-rakiewicz M, Mahmud N, Guy M, O’Brien LT, Wilkinson RA,Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Hor-wich A, Huddart RA, Khoo VS, Parker CC, Van As N, Wood-house CJ, Thompson A, Christmas T, Ogden C, Cooper CS,Lophatonanon A, Southey MC, Hopper JL, English DR, WahlforsT, Tammela TL, Klarskov P, Nordestgaard BG, Roder MA, Tybj-aerg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wi-klund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC,Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitz-gerald L, Kolb S et al (2011) Seven prostate cancer susceptibilityloci identiWed by a multi-stage genome-wide association study.Nat Genet 43:785–791. doi:10.1038/ng.882
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Kos-kenvuo M, Pukkala E, Skytthe A, Hemminki K (2000) Environ-mental and heritable factors in the causation of cancer–analyses ofcohorts of twins from Sweden, Denmark, and Finland. N Engl JMed 343:78–85. doi:10.1056/NEJM200007133430201
Lindstrom S, Adami HO, Adolfsson J, Wiklund F (2008) Y chromo-some haplotypes and prostate cancer in Sweden. Clin Cancer Res14:6712–6716. doi:10.1158/1078-0432.CCR-08-0658
Ma J, Li H, Giovannucci E, Mucci L, Qiu W, Nguyen PL, Gaziano JM,Pollak M, Stampfer MJ (2008) Prediagnostic body-mass index,plasma C-peptide concentration, and prostate cancer-speciWc mor-tality in men with prostate cancer: a long-term survival analysis.Lancet Oncol 9:1039–1047. doi:10.1016/S1470-2045(08)70235-3
Paracchini S, Pearce CL, Kolonel LN, Altshuler D, Henderson BE,Tyler-Smith C (2003) A Y chromosomal inXuence on prostatecancer risk: the multi-ethnic cohort study. J Med Genet 40:815–819
Rootsi S, Zhivotovsky LA, Baldovic M, Kayser M, Kutuev IA, Khu-sainova R, Bermisheva MA, Gubina M, Fedorova SA, IlumaeAM, Khusnutdinova EK, Voevoda MI, Osipova LP, StonekingM, Lin AA, Ferak V, Parik J, Kivisild T, Underhill PA, VillemsR (2007) A counter-clockwise northern route of the Y-chromo-some haplogroup N from Southeast Asia towards Europe. Eur JHum Genet 15:204–211. doi:10.1038/sj.ejhg.5201748
Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, WaterstonRH, Wilson RK, Page DC (2003) Abundant gene conversion be-tween arms of palindromes in human and ape Y chromosomes.Nature 423:873–876. doi:10.1038/nature01723
Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, LindstromS, Stevens VL, Chen C, Mondul AM, Travis RC, Stram DO, EelesRA, Easton DF, Giles G, Hopper JL, Neal DE, Hamdy FC, Dono-van JL, Muir K, Al Olama AA, Kote-Jarai Z, Guy M, Severi G,Gronberg H, Isaacs WB, Karlsson R, Wiklund F, Xu J, Allen NE,Andriole GL, Barricarte A, Boeing H, Bas Bueno-de-Mesquita H,
123
http://dx.doi.org/ng1999[pii]10.1038/ng1999http://dx.doi.org/ng1999[pii]10.1038/ng1999http://dx.doi.org/10.1038/ng2062http://dx.doi.org/10.1038/ng2062http://dx.doi.org/10.1038/ng.89http://dx.doi.org/10.1038/ng.448http://dx.doi.org/10.1038/ng2015http://dx.doi.org/10.1007/s00439-009-0727-5http://dx.doi.org/10.1007/s00439-009-0727-5http://dx.doi.org/10.1101/gr.7172008http://dx.doi.org/10.1371/journal.pone.0000172http://dx.doi.org/10.1038/ng.882http://dx.doi.org/10.1056/NEJM200007133430201http://dx.doi.org/10.1158/1078-0432.CCR-08-0658http://dx.doi.org/10.1016/S1470-2045(08)70235-3http://dx.doi.org/10.1038/sj.ejhg.5201748http://dx.doi.org/10.1038/nature01723
-
Hum Genet (2012) 131:1173–1185 1185
Crawford ED, Diver WR, Gonzalez CA, Gaziano JM, Gio-vannucci EL, Johansson M, Le Marchand L, Ma J, Sieri S, StattinP, Stampfer MJ, Tjonneland A, Vineis P, Virtamo J, Vogel U,Weinstein SJ, Yeager M, Thun MJ, Kolonel LN, Henderson BE,Albanes D, Hayes RB, Spencer Feigelson H, Riboli E, Hunter DJ,Chanock SJ, Haiman CA, Kraft P (2011) Genome-wide associa-tion study identiWes new prostate cancer susceptibility loci. HumMol Genet 20:3867–3875. doi:10.1093/hmg/ddr295
Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V,Maccioni L, Triantaphyllidis C, Shen P, Oefner PJ, ZhivotovskyLA, King R, Torroni A, Cavalli-Sforza LL, Underhill PA, Sant-achiara-Benerecetti AS (2004) Origin, diVusion, and diVerentia-tion of Y-chromosome haplogroups E and J: inferences on theneolithization of Europe and later migratory events in the Medi-terranean area. Am J Hum Genet 74:1023–1034. doi:10.1086/386295
Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, KawaguchiT, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Na-kamura Y, Nakagawa H (2010) Genome-wide association studyidentiWes Wve new susceptibility loci for prostate cancer in theJapanese population. Nat Genet 42:751–754. doi:10.1038/ng.635
The ATBC Cancer Prevention Study Group (1994) The alpha-tocoph-erol, beta-carotene lung cancer prevention study: design, meth-ods, participant characteristics, and compliance. The ATBCCancer Prevention Study Group. Ann Epidemiol 4:1–10
Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K,Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tas-sin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X,Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Al-banes D, Virtamo J, Weinstein S, Schumacher FR, GiovannucciE, Willett WC, Cussenot O, Valeri A, Andriole GL, CrawfordED, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoover R, HayesRB, Hunter DJ, Chanock SJ (2008) Multiple loci identiWed in agenome-wide association study of prostate cancer. Nat Genet40:310–315. doi:10.1038/ng.91
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, FoleyRA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography ofY chromosome binary haplotypes and the origins of modernhuman populations. Ann Hum Genet 65:43–62. pii:S0003480001008582
Valeri A, Briollais L, Azzouzi R, Fournier G, Mangin P, Berthon P,Cussenot O, Demenais F (2003) Segregation analysis of prostate
cancer in France: evidence for autosomal dominant inheritanceand residual brother–brother dependence. Ann Hum Genet67:125–137. pii:022
Vijayakumar S, Garcia D, Hensel CH, Banerjee M, Bracht T, Xiang R,Kagan J, Naylor SL (2005) The human Y chromosome suppressesthe tumorigenicity of PC-3, a human prostate cancer cell line, inathymic nude mice. Genes Chromosomes Cancer 44:365–372.doi:10.1002/gcc.20250
Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N(2004) Assessing the probability that a positive report is false: anapproach for molecular epidemiology studies. J Natl Cancer Inst96:434–442
Wellcome Trust Case Control Consortium (2007) Genome-wide associ-ation study of 14,000 cases of seven common diseases and 3,000shared controls. Nature 447:661–678. doi:10.1038/nature05911
Wiik K (2008) Where did European men come from? J Gen Genealogy4:35–85
Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Mini-chiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R,Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Al-banes D, Virtamo J, Weinstein S, Schumacher FR, GiovannucciE, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, AndrioleGL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF Jr, Hoo-ver R, Hunter DJ, Chanock SJ, Thomas G (2007) Genome-wideassociation study of prostate cancer identiWes a second risk locusat 8q24. Nat Genet 39:645–649. doi:10.1038/ng2022
Y Chromosome Consortium (2002) A nomenclature system for thetree of human Y-chromosomal binary haplogroups. Genome Res12:339–48. doi:10.1101/gr.217602
Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J,Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, Yu K, Hutch-inson A, Wang Z, Amundadottir L, Feigelson HS, Thun MJ, Div-er WR, Albanes D, Virtamo J, Weinstein S, Schumacher FR,Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, CrawfordED, Haiman CA, Henderson B, Kolonel L, Le Marchand L, Sid-diq A, Riboli E, Key TJ, Kaaks R, Isaacs W, Isaacs S, Wiley KE,Gronberg H, Wiklund F, Stattin P, Xu J, Zheng SL, Sun J, VattenLJ, Hveem K, Kumle M, Tucker M, Gerhard DS, Hoover RN,Fraumeni JF Jr, Hunter DJ, Thomas G, Chanock SJ (2009) Iden-tiWcation of a new prostate cancer susceptibility locus on chromo-some 8q24. Nat Genet 41:1055–1057. doi:10.1038/ng.444
123
http://dx.doi.org/10.1093/hmg/ddr295http://dx.doi.org/10.1086/386295http://dx.doi.org/10.1086/386295http://dx.doi.org/10.1038/ng.635http://dx.doi.org/10.1038/ng.91http://dx.doi.org/10.1002/gcc.20250http://dx.doi.org/10.1038/nature05911http://dx.doi.org/10.1038/ng2022http://dx.doi.org/10.1101/gr.217602http://dx.doi.org/10.1038/ng.444
Y chromosome haplogroups and prostate cancer in populations of European and Ashkenazi Jewish ancestryAbstractIntroductionResultsStage I association analysisPopulation substructure of E1b1b1c carriersLimited evidence for association to prostate cancer in Stage II analysisHaplogroup frequency and population distribution
DiscussionMaterials and methodsStudy populationMarker selection and genotypingStatistical analysisValidation by sequencing
URLsAcknowledgmentsReferences