genesdev.cshlp.orggenesdev.cshlp.org/.../Supplemental_Information.docx · Web viewthat display a...
Transcript of genesdev.cshlp.orggenesdev.cshlp.org/.../Supplemental_Information.docx · Web viewthat display a...

Supplemental Information
Inventory of Supplemental Information:
Supplemental Data:
Figure S1, related to Figure 1
Figure S2, related to Figure 2
Figure S3, related to Figure 2 and 4
Figure S4, related to Figure 5
Table S1-8
Supplemental Data
Supplemental Figure 1.
(A). Transgene siRNAs are predominantly 22 nt in length and have a 5’G, suggesting that they are
products of RNA-dependent RNA polymerases, that is, secondary siRNAs. (B). siRNAs mapping to rol-
6::lin-14 in the eri-6 mutant, as compared to wild type. (C). siRNAs mapping to sur-5::gfp in the eri-6
mutant, as compared to wild type. (D). sense transgene siRNAs are not depleted in eri-6 mutants. (E).
siRNAs mapping to the lin-14 3’UTR, present on the mgIs30 transgene. Indicated are the regions where
most microRNAs bind and the poly(A) signal. (F). Biological replicate of Figure 1E. (G). Biological
replicate of Figure 1F..


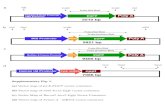
Supplemental Figure 2.
(A). The mgIs30 transgene is silenced upon gene inactivation of the eri-6/7 helicase gene by RNAi, no
longer displaying a rolling (Rol) phenotype. (B). Strategy used to identify genes that are required for
transgene expression. (C) Percentage of animals on a plate that display a silenced transgene phenotype,
averaged from six tests, for each of the 69 gene inactivations that score above the cut-off of 35%. Vector
control RNAi results in 2% of the animals on average displaying a silenced transgene phenotype (n=438
plates). (D). The 69 genes form interacting networks. (E). The 69 newly identified genes are enriched for
germline expression. (F). Motif identified in cluster 2 genes.


Supplemental Figure 3.
(A). Strategy to identify negative regulators and competing pathways of exogenous RNAi using gene
inactivation by RNAi. (B). siR-1 reads per million as determined by small RNA deep sequencing of wild
type C. elegans raised at 15, 20 and 25C. (C). Structure of the eri-12 gene, the dsRNA that targets eri-12
and mutations in this gene, cxTi7130.


Supplemental Figure 4.
(A). Sense siRNA reads are not depleted in the arl-8 mutant. (B). ERGO-1-ERI-6/7 target siRNAs are
reduced in the arl-8 mutant. (C). Reduced levels of ALG-3/4 target siRNAs in the eri-6/7 mutant. (D).
Reduced levels of a subset of ALG-3/4 target siRNAs (indicated by the purple oval) in the arl-8 mutant.
(E). arl-8 is required for ALG-3/4 target genes that are not CSR-1 targets. (F). Overlap between genes
transcriptionally co-regulated with eri-7/helicase, arl-8/Arf-like GTPase and F37A4.1.


Supplemental Table 1.Transgenes used in this study.
transgene promoter gene3'UTR
copy number tissue expressed
mirWIP score reference
chromosome
Prol-6::rol-6(su1006)::lin-14 3’UTR
rol-6 (collagen)
rol-6 (gof)
lin-14
high copy hypodermis 327.1
Grishok et al 2001 IV
mgIs30
Prol-6::rol-6(su1006)::rol-6 3’UTR
rol-6 (collagen)
rol-6 (gof)
rol-6
low copy hypodermis 19.55
Fischer et al 2008; Mello et al 1991
pkIs1582
Plet-858::let-858::gfp::let-858 3’UTR
let-858 (splicing factor)
let-858::gfp
let-858
low copy ubiquitous 2
Fischer et al 2008; Kelly et al 1997
pkIs1582
Psur-5::sur-5::gfp::unc-54 3’UTR
sur-5 (AA-CoA synthetase) gfp
unc-54
high copy
ubiquitous (scored intestine) 34.8
Yochem et al 1998
Pscm::gfp scm (synthetic)high copy hypodermal seam cells
Koh and Rothman 2001 V
wIs54
Pmyo-3::gfp::let-858 3’UTR
myo-3 (myosin heavy chain)
let-858
high copy body wall muscle 2
Winston et al 2002 I
ccIs4251
Phsp-4::gfp::unc-54 3’UTR hsp-4 (hsp70) gfp
unc-54
high copy
intestine, hypodermis, spermatheca 34.8
Calfon et al 2002 V
zcIs4
Supplemental Table 2. Silencing phenotypes.
Gene NameSequence
Name % silencing
rol-6(su1006)::rol-6 3'UTR
let-858::gfp::let-858 3'UTR
sur-5::gfp::unc-54 3'UTR scm::gfp
myo-3::gfp::let-858 3'UTR
hsp-4::gfp::unc-54 3'UTR
nrde-3::gfp shift
RNA binding/translation
puf-8 C30G12.7 38 silencedcytoplasmic
ain-1 C06G1.4 43 N/A silenced silenced nuclear
csr-1 F20D12.1 91 silenced silenced nuclear
vig-1 F56D12.5 39
ergo-1 R09A1.1 98 silenced silenced silenced silenced silencedcytoplasmic
rde-4 T20G5.11 93 silenced silenced silenced silenced silencedcytoplasmic
nuclear transport/transcription/RNA turnover
ntl-9 C26E6.3 76 N/A silenced silenced desil nuclear
lin-35 C32F10.2 65 silenced silenced silenced silenced silencedcytoplasmic
C35A5.8 C35A5.8 68 silenced
nurf-1 F26H11.2 36 silenced silenced

F29A7.6 F29A7.6 66 N/A silenced silenced silenced silenced
isw-1 F37A4.8 88 silenced silenced silenced nuclear
mes-3 F54C1.3 41 silenced
cpar-1 F54C8.2 43
lin-54 JC8.6 93 silenced silenced silenced silencedcytoplasmic
rnp-5 K02F3.11 98 desil silenced silenced silenced desil
mes-2 R06A4.7 62 desil silenced silenced silenced silenced nuclear
htz-1 R08C7.3 85 desil
R186.7 R186.7 53 silenced silenced
dpy-20* T22B3.1 85 silenced silenced
npp-12 T23H2.1 43 silenced
T26A8.4 T26A8.4 79 silenced silenced silenced nuclear
ell-1 Y24D9A.1 85 silenced N/A silenced silenced
exos-1 Y48A6B.5 48 desil
ccr-4 ZC518.3 36 desil
tcer-1 ZK1127.9 58 silenced desilcytoplasmic
lin-9 ZK637.7 93 silenced silenced silenced silenced silenced n/a
dpy-30 ZK863.6 48 silenced
attf-3 F54C4.3 73 silenced desil silenced silenced silenced silenced
other RNA or DNA binding proteins
eri-6 C41D11.1 93 silenced silenced silenced silenced silencedcytoplasmic
eri-7 C41D11.7 98 silenced silenced silenced silenced silencedcytoplasmic
drh-3 D2005.5 58 silenced silenced silenced silencedcytoplasmic
F58G11.2 F58G11.2 89
nucleic acid/protein modification
sams-3/-4C06E7.1/.3 60 desil desil nuclear
adr-1** H15N14.1 98 silenced N/A silencedcytoplasmic
D2005.1** D2005.1 98 silenced N/A silencedcytoplasmic
adr-2 T20H4.4 88 silenced silenced silenced
prmt-1Y113G7B.17 68 silenced silenced
cytoplasmic
protein folding/turnover
adt-2* F08C6.1 89 silenced Ste nuclear
ubc-20 F40G9.3 42 silencedcytoplasmic
stc-1 F54C9.2 77 Sck desil
suro-1* R11A5.7 77 silenced silenced

T27A3.2 T27A3.2 70 silenced desil silenced silenced
metabolic enzymes
mel-32C05D11.11 49 silenced silenced
cytoplasmic
pdi-2 C07A12.4 69 silenced desil
sdha-2 C34B2.7 62 silencedcytoplasmic
F37A4.1 F37A4.1 39 silenced silenced
hgo-1 W06D4.1 38 silenced silenced silenced desil silencedcytoplasmic
Y48A6B.7 Y48A6B.7 43 silenced
vesicular transport
cogc-5 C43E11.1 43 silenced silenced silenced silencedcytoplasmic
aps-2 F02E8.3 48 silenced silenced desil
dpy-23 R160.1 98 silenced silenced silenced
apa-2 T20B5.1 36 silenced Ste
arl-8Y57G11C.13 54 silenced silenced silenced
other
flna-1 C23F12.1 64 N/A silenced silenced
col-90* C29E4.1 56 silenced N/A desil desil
cutl-20* F54A5.2 39 silenced silenced nuclear
F35E8.4 F35E8.4 44 silenced
T11F9.7 T11F9.7 49 silenced silenced
ekl-1 F22D6.6 70 silenced silenced silenced silenced nuclear
ent-1 ZK809.4 54 silenced N/A silencedcytoplasmic
unknown
B0001.6 B0001.6 85 silenced silenced silencedcytoplasmic
C25F9.11 C25F9.11 36 silenced silenced nuclear
C41D11.4 C41D11.4 38 silenced silenced
C43H6.4 C43H6.4 45 silenced silenced silenced
F57C9.7 F57C9.7 48 silenced
T03F6.4 T03F6.4 43 silenced silencedcytoplasmic
T24C4.2 T24C4.2 45
Y39G10AR.7Y39G10AR.7 66 silenced silenced silenced

Supplemental Table 3. eri and rde genes identified- indicates no response to RNAi, + indicates a reponse to RNAi, ++ indicates a strong response to RNAi (stronger than loss of function mutant)
Double RNAi
Rde tester Mutant
Rde tester
Gene tested hmr-1 unc-73 lin-1 dpy-13 mom-2 hmr-1
unc-73
lin-1
dpy-13 pos-1 allele tested
eri genes arl-8 +/- +/- + ++ n/a + (soma) - - ++ n/a tm2472
B0001.6+ (soma only) - - + n/a + + + n/a + cxTi7130
C26E6.3 + - + + + + - + + + ok1728
ccr-4 + + +/- + n/a - - + + tm1312
D2005.1+ (soma only) - +/- - n/a + (soma) +/- - - + ok2689
ell-1+ (soma only) - - ++ n/a
ergo-1 + + + ++ n/a + + + ++ +tm1860, mg394
eri-7 + + + ++ + + + + ++ + mg369
F37A4.1+ (soma only) +/- + +/- + - - +/- n/a + ttTi2621
lin-35+ (soma only) + + n/a + + - + n/a + n745
lin-54 + - n/a ++ n/aWang et al (2005)
lin-9 + + n/a ++ n/aWang et al (2005)
mel-32+ (soma only) +/- n/a n/a n/a
mes-3 + + - +/- n/a
npp-12 - - - + n/a +/- + +/- ++ +ok2424, tm2320
puf-8 + - - ++ n/a + - + n/a + ok302
T11F9.7+ (soma only) - +/- ++ +
T24C4.2+ (soma only) + +/- n/a +
T26A8.4+ (soma only) - + + +
tcer-1 + - +/- ++ n/a
ubc-20 - +/- + - n/a
vig-1 n/a - +/- +/- n/a + (soma) + +/- ++ +tm3383, ok2536
wild type - - - + + - - - + +
rde genes rde-4 + - +/- n/a - n/a -
n/a n/a - ne299
csr-1 n/a + + - n/a n/aClaycomb et al (2009)
drh-3 n/a +/- + +/- n/a n/aDuchaine et al (2006)
ekl-1 - - - + -Rocheleau et al. (2008)

F58G11.2 - - - - n/a
htz-1 - +/- + +/- -
-: no silencing, +: moderate silencing, ++: strong silencing, +++: super silencing
Supplemental Table 4. Genes found most frequently coexpressed with genes identified in the transgene silencing screen
WB gene name
Gene Public Name
number of hits this gene is coexpressed with
Coexpressed with cluster Description
WBGene00002073 ima-2 12 1 importin alphaWBGene00007109 B0035.6 12 1WBGene00008385 D1081.7 11 1WBGene00011464 T05B9.1 11 1WBGene00018948 F56C9.3 10 1WBGene00003865 oma-2 9 1 zinc fingerWBGene00006311 sun-1 9 1
SUN domain; binds lamins on outer nuclear membrane
WBGene00008005 C38D4.4 9 1WBGene00010053 F54D5.9 9 1 F box proteinWBGene00018950 F56C9.6 9 1WBGene00000935 daz-1 8 1
RNA recog motif; req. for progression of meiosis during oogenesis
WBGene00002034 htp-3 8 1 him-3 paralog; req for chr segregationWBGene00004044 plk-3 8 1 polo-like kinaseWBGene00009270 F30F8.1 8 1WBGene00015504 C06A5.6 8 1WBGene00020035 egg-5 8 1 pseudotyrosine phosphataseWBGene00001514 xnd-1 7 1 X chromosome non-disjunction factorWBGene00003231 mex-6 7 1 CCCH zinc-fingerWBGene00004239 puf-3 7 1 pumilioWBGene00004244 puf-8 7 1 pumilioWBGene00007624 hrd-1 7 1 hrd-1/wago-9 ArgonauteWBGene00007762 C27B7.5 7 1 zinc fingerWBGene00010351 cbd-1 7 1 12 chitin-binding peritrophin-A domainsWBGene00011061 wago-1 7 1 ArgonauteWBGene00011489 T05F1.2 7 1WBGene00011886 T21B10.4 7 1WBGene00013729 Y111B2A.3 7 1 paralog of B0035.6WBGene00019002 F57B10.4 7 1WBGene00019608 ani-2 7 1 anillinWBGene00019833 R02F2.4 7 1 6 chitin-binding peritrophin-A domains

WBGene00020948 W02F12.3 7 1WBGene00003158 mcm-6 15 2 DNA replication licensing factorWBGene00003036 lin-53 13 2 DRM complexWBGene00003183 mei-1 13 2 meiotic spindle formation, kataninWBGene00000913 daf-18 12 2 PTENWBGene00006474 wdr-5.1 12 2 H3K4 trimethylation complexWBGene00006773 unc-37 11 2
WD-repeat protein; interacts with Mediator complex
WBGene00009254 capg-1 11 2 condensin I complexWBGene00001102 dsh-2 10 2WBGene00004392 rnr-2 10 2 ribonucleotide reductaseWBGene00007979 imp-1 10 2 intramembrane proteaseWBGene00011636 cec-3 10 2 chromodomainWBGene00017546 rpa-1 10 2 RPA large subunit; dna replication, repairWBGene00000411 cdl-1 9 2 stemloop binding in histone 3'UTRWBGene00000875 cyk-4 9 2 Rho GAP; enriched in spermWBGene00001259 emb-5 9 2 Spt6 RNA pol II TFWBGene00001827 hcf-1 9 2
transcriptional regulator that associates with histone modification enzymes
WBGene00001834 hda-1 9 2 histone deacetylase 1WBGene00003157 mcm-5 9 2 DNA replication licensing factorWBGene00007110 B0035.11 9 2 RNA-polymerase-associated protein LEO1WBGene00008386 D1081.8 9 2 cell division cycle 5WBGene00011625 vps-39 9 2 vacuolar protein sortingWBGene00011747 sna-2 9 2 snRNP-binding proteinWBGene00016803 bath-42 9 2 BTB and MATH domainWBGene00044916 F40F8.11 9 2WBGene00000794 crn-1 8 2 5'3'exonuclease cell death relatedWBGene00000868 cyb-3 8 2 cyclin B; req for chr segregationWBGene00001568 gei-11 8 2 Myb DNA binding domainWBGene00001874 him-17 8 2 required for H3K9 demethylationWBGene00002027 hsr-9 8 2 BRCT domain; DNA damage responseWBGene00003159 mcm-7 8 2 DNA replication licensing factorWBGene00003792 npp-6 8 2 nucleoporinWBGene00004204 swsn-1 8 2 swi/snfWBGene00006776 unc-40 8 2 netrin receptorWBGene00006974 zen-4 8 2 kinesin-like proteinWBGene00007042 pbrm-1 8 2 polybromo domainWBGene00007403 set-3 8 2 SET domainWBGene00009661 patr-1 8 2 decapping activator; required for P bodiesWBGene00017085 E01A2.2 8 2 SerrateWBGene00017643 czw-1 8 2 centromere/kinetochore protein zw10WBGene00000271 brf-1 7 2 TATA bindingWBGene00001584 nrde-4 7 2 nuclear RNAi deficientWBGene00001743 grp-1 7 2 GTP exchange factor for ARFsWBGene00003406 mrg-1 7 2 nuclear; localizes to autosomesWBGene00003472 mtk-1 7 2 map kinase kinase kinase; JK screenWBGene00003576 ndc-80 7 2 COG-7WBGene00003788 npp-2 7 2 nucleoporinWBGene00003805 npp-19 7 2 nucleoporin

WBGene00003955 pcn-1 7 2 PCNA DNA replication and repairWBGene00004874 smc-4 7 2 subunit of mitotic condensinWBGene00007256 swsn-9 7 2 bromodomainWBGene00007433 swsn-7 7 2 swi/snf; zn finger domainWBGene00008035 C39E9.12 7 2 SAP domainWBGene00009922 F52B5.3 7 2 DEAH helicase; SplindleEWBGene00011758 T13H5.4 7 2 splicing factor 3a subunit 3; PRP9-likeWBGene00015017 rpl-2 7 2 large ribosomal protein L8WBGene00016154 C27A12.2 7 2 zinc-finger proteinWBGene00016238 C30A5.3 7 2 CCR4 complex memberWBGene00020111 R151.8 7 2WBGene00020821 jhdm-1 7 2 histone demethylaseWBGene00017816 F26B1.2 10 3 K homology domainWBGene00019893 sgt-1 9 3 glutamine-rich tetratrico repeat proteinWBGene00004980 spk-1 8 3 SR protein kinaseWBGene00020184 T03F1.1 8 3 UBA5WBGene00004462 rpn-6.1 7 3 subunit of 19S regulatory complexWBGene00006528 tba-1 7 3 alpha-tubulinWBGene00006529 tba-2 7 3 alpha-tubulinWBGene00007588 C14C10.5 7 3 proteasome activator complexWBGene00003247 mig-15 11 4 Nck-interacting kinaseWBGene00000482 chd-3 9 4 NuRDWBGene00004951 spc-1 8 4 alpha spectrin orthologWBGene00017423 F13C5.2 8 4 bromodomain containing; nurf-1 paralogWBGene00004140 pqn-55 7 4 zinc-finger SWIM typeWBGene00004148 pqn-65 7 4WBGene00018520 F46H5.4 7 4 cytokinesis proteinWBGene00018710 F52G3.1 7 4WBGene00018743 F53B3.5 7 4 claudin homologWBGene00017132 mel-47 10 1, 2 RNA recognition motif; tiar-1 paralogWBGene00011560 T07C4.3 9 1, 2WBGene00007978 C36B1.11 8 1, 2WBGene00002068 ify-1 7 1, 2WBGene00017317 attf-2 17 2, 3 AT hook transcription factorWBGene00018967 F56D2.6 11 2, 3 DEAH helicase; pre-mRNA splicingWBGene00011605 T08A11.2 10 2, 3 splicing factor 3b subunit 1 (SAP155)WBGene00007105 B0035.1 9 2, 3 Zn-finger C2H2-likeWBGene00000836 cul-1 8 2, 3 cullinWBGene00003085 lst-3 8 2, 3 transcription factor; SynMuvWBGene00001999 hrp-1 7 2, 3 hnRNP required for mRNA surveillanceWBGene00003795 npp-9 7 2, 3 nucleoporinWBGene00004187 prp-8 7 2, 3 pre-mRNA splicing factorWBGene00006856 usp-14 7 2, 3 ubiquitin-specific proteaseWBGene00019323 teg-4 7 2, 3 pre-mRNA splicing factorWBGene00020423 prp-19 7 2, 3 splicing factor; wdr-5.1 paralogWBGene00018849 F55A3.3 14 2, ccr-
4/ntl-9 FACT complex; associates chromatinWBGene00015943 tiar-1 9 3, 2 TIA-1/TIAL RNA binding protein homolog

Supplemental Table 5. Strains used in this study.Strain name Gene name allele nameCB1282 dpy-20 e1282CB840 dpy-23 e840DH1230 chc-1 b1025EG2988 dpy-23 gm17EG4015 dpy-23 rescuedFX1312 ccr-4 tm1312FX1420 C34B2.7 tm1420FX1452 ZK1127.9 tm1452FX1860 R09A1.1 tm1860FX2320 npp-12 tm2320FX2472 arl-8 tm2472 4xFX2472 arl-8 tm2472FX3383 vig-1 tm3383GR1200 sqt-1 sc13GR1702 sur-5::gfp
GR1703 lim-6::gfp, col-10::lacZ::lin-14, rol-6(su1006)::lin-14 3’-UTR
GR1704 eri-6 mg379HC57 myo-2::gfp; myo-3::gfpHE1006 rol-6 su1006JH1521 puf-8 ok302JR672 scm::gfpMT111 lin-8 n111MT13664 nurf-1 n4293/balMT15795 isw-1 n3294MT1806 lin-15 n767MT2131 lin-31 n1053MT301 lin-31 n301MT8677 ksr-1 n2526N2 wtRB1477 C26E6.3 ok1728RB1554 R186.7 ok1872RB1648 F54C4.3 ok2037RB1849 C43E11.11 ok2393RB1874 npp-12 ok2424RB1933 vig-1 ok2536RB2031 D2005.1 ok2689RB2047 pmrt-1 ok2710RB2165 C06E7.1RB891 ent-1SJ4005 hsp-4::gfpTY1936 dpy-30/balVC1196 K02F3.10&rnp-5 ok1634VC1270 lin-31 gk569

VC593 Y48A6B.5 ok807WM158 ergo-1 tm1860ZT3 csr-1 bal
B0001.6 cxTi7130pkIs1582 (let-858::gfp (low copy);rol-6)C41D11.1 ttTi6525C43H6.4 cxTi10129ekl-1 ttTi4868eri-1 mg366eri-7 mg411F37A4.1 ttTi2621F49F1.11 cxP6306lin-35 n745rde-1 ne300rde-4 ne299rrf-3 pk1426
Supplemental Table 6. Primers used in this study.gene sequenceZK380.5 forward agattgtctgcgacatgcacZK380.5 reverse gacatggtggcacttgaatgT08B6.2 forward cccgaacaaacggaatatgtT08B6.2 reverse gagtgacccgagaccagaaaF39E9.7 forward cccagtggcccaattaaacgF39E9.7 reverse cccacggcttgttctttgacaF55C9.3 forward ggaaaaccggaatcattcaaF55C9.3 reverse agctccacgttgtagcgtctC36A4.11 forward aaggcgaagaacgacagagaC36A4.11 reverse tccaattcgaaggcaatcttlin-14 forward ataaggcccaaagtggaaaclin-14 reverse aaactttcaagctggctctttaeri-6/7 forward cgtcgtcctattgggaaatgeri-6/7 reverse tggaatcgaacgaattgagcY45F10D.4 forward gtcgcttcaaatcagttcagcY45F10D.4 reverse gttcttgtcaagtgatccgacarpl-32 forward caaggtcgtcaagaagaagcrpl-32 reverse ggctacacgacggtatctgt

Supplemental Table 7. Taqman assays used in this study.Applied BiosystemsGene Target Sequence Assay Name Assay IDC40A11.10 (Han et al 2009)
GCUCAGAAACGGUAGAUUAUUUUCAA siR26-O1 CCY892P
E01G4.7 (Han et al 2009)
GUUAUGACUCAUAAUUGCAGCAAAUG siR26-O2 CCX0BWH
siR-1 (A) GAAUAGAUACGCGGUAUGAG xcluster-A CSMSF7GsiR-1 (B) GAAUAGAUACGCGGUAUGAGGU xcluster-B CSN1EDOE01G4.5 GUCACAAACUACGUCUUUUUCU E01G4.522G CS0IV5Rmir-35 UCACCGGGUGGAAACUAGCAGU cel-miR-35 196mir-1 UGGAAUGUAAAGAAGUAUGUA has-miR-1 385lin-4 UCCCUGAGACCUCAAGUGUGA cel-lin-4 258
lin-14 3’UTR GTACAATGGGATGGATGTTAAGA
S. c. Human C. elegans Description Tested? Screen Retest
Caf40 CNOT9/RQCD1 ntl-9 ✓ strong +Ccr4 CCR4/ CNOT6 ccr-4 deadenylase ✓ strong +Not2 CNOT2 ntl-2/B0286.4 ✓ strong + LvaNot4 CNOT4 ntl-4/C49H3.5 E3 ligase ✓ med. + did not
retestCaf1 CNOT7,CNOT8 ccf-1/Y56A3A.20 deadenylase ✓ EmbNot1 CNOT1 ntl-1/F57B9.2 ✓ LetNot3 CNOT3 ntl-3/Y56A3A.1 ✓ EmbNot5 CNOT5 ntl-3/Y56A3A.1 ✓ Emb
Supplemental Table 8. CCR4/NOT complex component gene inactivations identified as transgene
silencers. S.c. stands for Saccheromyces cerevisiae. Emb= embryonic lethal, Let= lethal, Lva = larval;
arrest. Screen shows the results from the initial genome wide screen.