Supplementary Figures and Tables A novel gene THSD7A is ... · Supplementary Figures and Tables A...
Transcript of Supplementary Figures and Tables A novel gene THSD7A is ... · Supplementary Figures and Tables A...

Supplementary Figures and Tables
A novel gene THSD7A is associated with obesity
S Nizamuddin, P Govindaraj, S Saxena, M Kashyap, A Mishra, S Singh, H Rotti, R Raval, J Nayak, BK Bhat, BV Prasanna, VR Dhumal, S Bhale, KS Joshi, AP Dedge, R Bharadwaj, GG Gangadharan, S Nair, PM Gopinath, B Patwardhan, P Kondaiah, K Satyamoorthy, MS Valiathan and K Thangaraj
Supplementary Figure S1 1
Supplementary Figure S2 2
Supplementary Figure S3 3
Supplementary Figure S4 4
Supplementary Figure S5 5
Supplementary Table S1 6
Supplementary Table S2 12
Supplementary Table S3 13
Supplementary Table S4 18
Supplementary Table S5 19
Supplementary Table S6 20
Supplementary Table S7 21

1
Supplementary Figure S1. Geographical location of 208 subjects used in
discovery stage. All subjects are from Indian sub-continent, except one is from
Nepal.

2
Supplementary Figure S2. Quantile-Quantile plot for the p-value (792659 SNPs)
achieved in genome-wide association study.

3
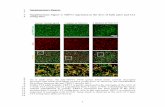
Supplementary Figure S3. Regional plot of rs11645605 (intronic SNP of WWOX) with BMI distribution for each
genotype. We considered allele codes on positive strand and represented ancestral alleles in italic.

4
Supplementary Figure S4. Genotyping of 4 SNPs associated with BMI in replication samples with Sanger sequencing
method. A) rs9551868: KATNAL1, B) rs2281524: MTF2, C) rs1526538: THSD7A and D) rs7681875: LIMCH1.

5
Supplementary Figure S5. Regional plot of THSD7A with ±10kb flanking region. The p-values were obtained from
GIANT database.

6
Supplementary Table S1. Sample details of Discovery stage (cohort-I) samples.
Sample-ID Nearest City State/Country Latitude Longitude Caste BMI Category SD049 Neerchal Kerala 12.588388 75.034829 Kasargod_Ujire:Hindu 14.01 Underweight BN121 Bangalore Karnataka 12.971599 77.594563 Hindu 15 Underweight PN17_1033 Bharsi Uttar Pradesh 29.29681 77.302287 Maratha:Hindu 15.4 Underweight BN033 Bangalore Karnataka 12.971599 77.594563 Hindu 15.6 Underweight PN2_055 Pune Maharastra 18.52043 73.856744 Sonar:Hindu 15.8 Underweight BN160 - Tamil Nadu 13.178318 80.249366 Catholic,Christian 16.1 Underweight PN14_653 Raipur Chattisgarh 21.251384 81.629641 Marwadi:Hindu 16.4 Underweight PN17_1023 Satara Maharastra 17.691401 74.000938 Maratha:Hindu 16.5 Underweight PN16_862 Sangli Maharastra 16.85438 74.564171 Maratha:Hindu 16.6 Underweight PN11_335 Solapur Maharastra 17.659919 75.906391 Bhavsar:Hindu 16.7 Underweight SD238 Kukkehalli Karnataka 13.376018 74.856561 Martti:Hindu 16.79 Underweight PN18_1191 Nashik Maharastra 19.997453 73.789802 Kunabi:Hindu 17 Underweight BN162 Mumbai Maharastra 19.075984 72.877656 Hindu 17.3 Underweight BN289 Bangalore Karnataka 12.971599 77.594563 Hindu 17.3 Underweight BN190 - Kerala 10.850516 76.271083 Hindu 17.6 Underweight PN18_1049 - Jammu and Kashmir 34.149088 76.825965 Bramhin:Hindu 17.6 Underweight PN18_1258 Kolhapur Maharastra 16.691308 74.244866 Dhanagar:Hindu 17.6 Underweight BN020 Bangalore Karnataka 12.971599 77.594563 Hindu 17.6 Underweight BN038 - Haryana 29.058776 76.085601 Hindu 17.7 Underweight BN061 - Andhra Pradesh 17.339609 78.43825 Hindu 17.7 Underweight PN7_214 Satara Maharastra 17.691401 74.000938 Maratha:Hindu 17.7 Underweight BN071 - Karnataka 15.317278 75.713888 Hindu 17.9 Underweight PN_1040 Pune Maharastra 18.52043 73.856744 Kunabi:Hindu 17.9 Underweight PN18_1128 Chalisgaon Maharastra 20.456808 75.003068 Mali:Hindu 17.9 Underweight SD247 Kalyanpur Uttar Pradesh 29.003426 79.397235 Padmashali:Hindu 17.95 Underweight SD114 Udupi Karnataka 13.33232 74.746048 Sherigar:Hindu 17.99 Underweight BN070 - Andhra Pradesh 17.339609 78.43825 Muslim 18 Underweight PN17_993 Ahmednagar Maharastra 19.095208 74.749592 Chambar:Hindu 18 Underweight BN120 Raichur Karnataka 16.20702 77.354362 Hindu 18.1 Underweight BN247 - Gujrat 23.021453 72.582146 Hindu 18.1 Underweight BN259 Mavalikkara Kerala 9.250576 76.540081 Konkani,Hindu 18.1 Underweight BN271 Bangalore Karnataka 12.971599 77.594563 Vokkaliga:Hindu 18.1 Underweight PN02_61 Pulghar Maharastra 19.701989 72.774733 Bramhin:Hindu 18.1 Underweight SD194 Gangavathi Karnataka 15.434481 76.530272 Moodibidri_Lingayat:Hindu 18.16 Underweight

7
PN12_452 Kopargaon Maharastra 19.8833 74.4833 Maratha:Hindu 18.3 Underweight BN107 - Uttar Pradesh 27.570589 80.098187 Shiya:Hindu 18.4 Underweight PN16_882 Pune Maharastra 18.52043 73.856744 Konkani,Hindu 18.4 Underweight PN6_183 Aurangabad Maharastra 19.876165 75.343314 Banjara:Hindu 18.4 Underweight BN119 Bidar Karnataka 17.92 77.519722 Hindu 18.5 Normal PN14_728 Nanded Maharastra 19.153061 77.305847 Bramhin:Hindu 18.5 Normal BN147 Hyderabad Telangana 17.385044 78.486671 Hindu 18.9 Normal PN18_1045 Pune Maharastra 18.52043 73.856744 Maratha:Hindu 18.9 Normal BN126 Tumkur Karnataka 13.338581 77.101219 Hindu 19 Normal PN18_1108 Kritsagar Bihar 25.096074 85.313119 Bramhin:Hindu 19 Normal PN18_1135 Chandrapur Maharastra 19.95 79.3 Mali:Hindu 19 Normal PN_1031 Nipani Karnataka 15.317278 75.713888 Sali:Hindu 19.1 Normal PN12_487 Ambegaon Maharastra 19.112316 73.730133 Maratha:Hindu 19.1 Normal PN13_606 Ahmednagar Maharastra 19.095208 74.749592 Maratha:Hindu 19.1 Normal PN18_1149 Solapur Maharastra 17.659919 75.906391 Maratha:Hindu 19.1 Normal BN290 Bangalore Karnataka 12.971599 77.594563 Sunni:Muslim 19.3 Normal SD048 Puttur Karnataka 13.43838 79.551914 Bramhin:Hindu 19.37 Normal BN019 Bangalore Karnataka 12.971599 77.594563 Hindu 19.4 Normal SD226 - Kerala 10.850516 76.271083 Christian 19.46 Normal PN18_1181 Wani Maharastra 20.05241 78.95639 Mahar:Hindu 19.5 Normal PN5_801 Nanded Maharastra 19.153061 77.305847 Jain:Hindu 19.5 Normal PN18_1105 Phaltan Maharastra 17.984451 74.436042 Bramhin:Hindu 19.7 Normal SD223 Mandya Karnataka 12.5242 76.8958 Roman catholic:Christian 19.71 Normal BN168 Bangalore Karnataka 12.971599 77.594563 Hindu 19.8 Normal PN18_1114 Malegaon Maharastra 20.549936 74.528751 Chambhar:Hindu 19.8 Normal SD202 Hoovina Hadagali Karnataka 15.019953 75.931808 Valmiki:Hindu 19.83 Normal SD146 Udupi Karnataka 13.33232 74.746048 Roman catholic:Christian 19.85 Normal BN285 Bangalore Karnataka 12.971599 77.594563 - 19.9 Normal BN169 Bangalore Karnataka 12.971599 77.594563 Muslim 20 Normal PN15_776 Nashik Maharastra 19.997453 73.789802 Vaisnav:Hindu 20 Normal SD222 Neelavara Karnataka 13.455577 74.771263 Devadiga:Hindu 20.02 Normal SD309 Kundapur Karnataka 21.701792 87.091142 Kodi_Kanyana_Mogavera:Hindu 20.06 Normal BN045 Calicut Kerala 11.258753 75.78041 Hindu 20.1 Normal BN135 Basavakalyan Karnataka 17.8667 76.95 Muslim 20.1 Normal BN244 Kolkata West Bengal 22.572646 88.363895 Hindu 20.1 Normal SD315 Hubli Karnataka 15.364708 75.123955 Pernankila_RSB:Hindu 20.19 Normal BN060 - Andhra Pradesh 17.339609 78.43825 Hindu 20.2 Normal BN083 - Uttar Pradesh 27.570589 80.098187 Bramhin:Hindu 20.2 Normal

8
BN089 - Karnataka 15.317278 75.713888 Lingayat:Hindu 20.2 Normal BN261 - Kerala 10.850516 76.271083 Hindu 20.2 Normal SD014 Kottayam Kerala 9.59872 76.528897 Bramhin:Hindu 20.2 Normal BN156 Mandya Karnataka 12.5242 76.8958 Hindu 20.3 Normal SD272 Udupi Karnataka 13.33232 74.746048 Mogavera:Hindu 20.31 Normal BN148 Bangalore Karnataka 12.971599 77.594563 Hindu 20.4 Normal BN099 - Kerala 10.850516 76.271083 Vaishya:Hindu 20.5 Normal BN191 - Kerala 10.850516 76.271083 Hindu 20.5 Normal BN081 - Andhra Pradesh 17.339609 78.43825 Hindu 20.6 Normal SD051 - Nepal 28.394857 84.124008 Koirala:Hindu 20.6 Normal BN056 - Andhra Pradesh 17.339609 78.43825 Hindu 20.7 Normal PN18_1101 Bhusawal Maharastra 21.047371 75.787659 Leva_Patil:Hindu 20.7 Normal PN18_1210 Nashik Maharastra 19.997453 73.789802 Rangari:Hindu 20.7 Normal SD288 Bangalore Karnataka 12.971599 77.594563 Gowda:Hindu 20.76 Normal SD303 Sasthan Karnataka 13.465536 74.713222 GSB:Hindu 20.76 Normal BN016 Bangalore Karnataka 12.971599 77.594563 Hindu 20.8 Normal BN149 Bangalore Karnataka 12.971599 77.594563 Gowda:Hindu 20.8 Normal SD190 Pune Maharastra 18.52043 73.856744 Moodibidri_Bramhin:Hindu 20.82 Normal PN11_421 Mumbai Maharastra 19.075984 72.877656 Mali:Hindu 20.9 Normal PN15_829 Solapur Maharastra 17.659919 75.906391 Bramhin:Hindu 21 Normal BN084 - Himachal Pradesh 31.110375 77.16904 Bramhin:Hindu 21.1 Normal BN279 - Kerala 10.850516 76.271083 Nair:Hindu 21.1 Normal PN18_1062 Sehore Madhya Pradesh 23.2 77.0833 Muslim 21.1 Normal BN140 Ramanagar Karnataka 12.720679 77.282648 Vokkaliga:Hindu 21.2 Normal BN062 - Andhra Pradesh 17.339609 78.43825 Hindu 21.3 Normal BN205 Gundlupet Karnataka 11.808081 76.691759 Hindu 21.3 Normal PN18_1234 Mumbai Maharastra 19.075984 72.877656 Maratha:Hindu 21.3 Normal PN18_1259 Majri Maharastra 30.724325 75.790198 Bhudda:Hindu 21.3 Normal SD181 Kakkayangadu Kerala 11.93594 75.710254 Moodibidri_Veluthedam:Hindu 21.35 Normal PN12_442 Mumbai Maharastra 19.075984 72.877656 Bramhin:Hindu 21.4 Normal PN18_1093 Kolkata West Bengal 22.572646 88.363895 Maratha:Hindu 21.4 Normal BN166 Bangalore Karnataka 12.971599 77.594563 Hindu 21.5 Normal BN210 Theni Tamil Nadu 10.010366 77.476811 Hindu 21.5 Normal BN212 Bangalore Karnataka 12.971599 77.594563 Hindu 21.5 Normal PN14_706 Pune Maharastra 18.52043 73.856744 Maratha:Hindu 21.5 Normal BN073 - Kerala 10.850516 76.271083 Hindu 21.6 Normal PN18_1159 Bhagalpur Bihar 25.24003 86.984512 Kurmi:Hindu 21.6 Normal SD206 Allahabad Uttar Pradesh 25.435801 81.846311 Hindu 21.63 Normal

9
SD161 Sangli Maharastra 16.85438 74.564171 Marati:Hindu 21.67 Normal BN097 - Jammu and Kashmir 34.149088 76.825965 Pandit:Hindu 21.7 Normal SD052 Guntur Andhra Pradesh 16.29851 80.433647 Bramhin:Hindu 21.79 Normal BN059 - Andhra Pradesh 17.339609 78.43825 Hindu 21.9 Normal BN004 Bangalore Karnataka 12.971599 77.594563 Hindu 22 Normal BN185 Nashik Maharastra 19.997453 73.789802 Hindu 22 Normal PN_1004 Solapur Maharastra 17.659919 75.906391 Maratha:Hindu 22 Normal PN17_1036 Pune Maharastra 18.52043 73.856744 Banjara:Hindu 22 Normal SD187 Udyavara Karnataka 13.309743 74.736633 Sherigar:Hindu 22.09 Normal BN041 - Uttar Pradesh 27.570589 80.098187 Hindu 22.1 Normal BN262 Gurgaon Haryana 28.459497 77.026638 Jain:Hindu 22.2 Normal PN14_707 Satara Maharastra 17.691401 74.000938 Maratha:Hindu 22.2 Normal SD210 Chikmanglore Karnataka 13.311821 75.770385 Muslim 22.22 Normal PN18_1083 - Haryana 28.061927 76.581377 Hatkar:Hindu 22.3 Normal SD193 Davangeri Karnataka 14.466344 75.92384 Moodibidri_Vishwakarma:Hindu 22.32 Normal SD165 Udyavara Karnataka 13.309743 74.736633 Billava:Hindu 22.38 Normal BN128 Bangalore Karnataka 12.971599 77.594563 Hindu 22.5 Normal BN186 - Kerala 10.850516 76.271083 Hindu 22.5 Normal PN15_766 Solapur Maharastra 17.659919 75.906391 Lingayat:Hindu 22.6 Normal BN076 - Jharkand 23.610181 85.279935 Hindu 22.7 Normal BN078 - Andhra Pradesh 17.339609 78.43825 Hindu 22.7 Normal SD056.P Udupi Karnataka 13.33232 74.746048 - 22.73 Normal BN115 Koppal Karnataka 15.536054 76.271083 Hindu 22.8 Normal BN0114 Harapanahalli Karnataka 14.786647 75.979791 Hindu 22.9 Normal BN065 Mumbai Maharastra 19.075984 72.877656 Hindu 22.9 Normal BN213 - Tamin Nadu 13.178318 80.249366 Hindu 22.9 Normal BN288 Bangalore Karnataka 12.971599 77.594563 Hindi 22.9 Normal PN13_535 Mumbai Maharastra 19.075984 72.877656 Christian 23 Overweight SD078 Vishakapatnam Andhra Pradesh 17.686816 83.218482 Bramhin:Hindu 23.03 Overweight BN024 Bangalore Karnataka 12.971599 77.594563 Christian 23.1 Overweight PN18_1048 Junnar Maharastra 19.20928 73.872589 Mali:Hindu 23.1 Overweight SD083 Udyavara Karnataka 13.309743 74.736633 Billava:Hindu 23.19 Overweight BN143 Mysore Karnataka 12.29581 76.639381 Lingayat:Hindu 23.3 Overweight BN277 Thrivanathapurum Kerala 8.487495 76.948623 Hindu 23.3 Overweight PN18_1099 Jalagaon Maharastra 21.013321 75.563972 Vani:Hindu 23.3 Overweight PN18_1115 Palthan Rajasthan 24.087454 74.856561 Maratha:Hindu 23.3 Overweight SD168 Davangeri Karnataka 14.466344 75.92384 Lingayat:Hindu 23.38 Overweight BN199 - Kerala 10.850516 76.271083 Muslim 23.4 Overweight

10
PN18_1183 Mumbai Maharastra 19.075984 72.877656 Bramhin:Hindu 23.4 Overweight BN055 - Karnataka 15.317278 75.713888 Hindu 23.5 Overweight BN157 Bangalore Karnataka 12.971599 77.594563 Gowda:Hindu 23.5 Overweight PN18_1111 Pune Maharastra 18.52043 73.856744 Bramhin:Hindu 23.5 Overweight PN18_1164 Pune Maharastra 18.52043 73.8567444 Nhavi:Hindu 23.5 Overweight PN13_542 Chakan Maharastra 18.760266 73.863035 Mali:Hindu 23.7 Overweight PN14_700 Wai Maharastra 17.948646 73.891704 Maratha:Hindu 23.7 Overweight SD070 Changanacherry Kerala 9.4667 76.55 Nair:Hindu 23.72 Overweight SD195 Calicut Kerala 11.258753 75.78041 Moodibidri_Nair:Hindu 23.78 Overweight PN13_607 Pune Maharastra 18.52043 73.856744 Maratha:Hindu 23.8 Overweight SD034 Sardarshahar Rajasthan 28.440419 74.493705 Indoriya:Hindu 23.83 Overweight SD040 Gudgeri Karnataka 15.12384 75.363403 Lingayat:Hindu 23.93 Overweight SD071 Kodihil Karnataka 12.962755 77.649317 Godalli:Hindu 23.93 Overweight PN18_1107 Nagpur Maharastra 21.1458 79.088155 Maheshwari:Hindu 24 Overweight BN280 - Kerala 10.850516 76.271083 Hindu 24.1 Overweight PN15_810 Sanganer Rajasthan 26.811201 75.783172 Bhavsar:Hindu 24.1 Overweight PN15_835 Shrirampur Maharastra 19.620297 74.654658 Marwadi:Hindu 24.1 Overweight PN2_018 - New Delhi 28.615608 77.209547 Jat:Hindu 24.1 Overweight BN018 Bangalore Karnataka 12.971599 77.594563 Hindu 24.2 Overweight BN063 - Kerala 10.850516 76.271083 Christian 24.2 Overweight BN074 - Bihar 25.096074 85.313119 Hindu 24.2 Overweight PN18_1142 Dahanu Maharastra 19.957793 72.745674 Bramhin:Hindu 24.3 Overweight SD012 Sakleshpura Karnataka 12.939735 75.785777 Vokkaliga:Hindu 24.48 Overweight BN281 - Kerala 10.850516 76.271083 Hindu 24.6 Overweight PN15_905 Jhodpur Rajasthan 26.238947 73.024309 Jain:Hindu 24.6 Overweight BN215 Bangalore Karnataka 12.971599 77.594563 Hindu 24.8 Overweight SD271 Udupi Karnataka 13.33232 74.746048 ScheduleCaste:Hindu 24.81 Overweight BN206 Bangalore Karnataka 12.971599 77.594563 Hindu 24.9 Overweight BN102 - Kerala 10.850516 76.271083 Shiya:Muslim 25.1 Obese PN18_1100 Dhule Maharastra 20.90422 74.774898 Bramhin:Hindu 25.1 Obese PN18_1087 Thane Maharastra 19.218331 72.97809 Kshatriya:Hindu 25.2 Obese SD020 Sunnari Maharastra 19.083865 72.90703 Bunt:Hindu 25.2 Obese SD104 Naragund Karnataka 15.71941 75.384354 ganiga:Hindu 25.39 Obese BN257 Attingal Kerala 8.695205 76.811256 Hindu 25.6 Obese PN_1073 Mumbai Maharastra 19.075984 72.877656 Bramhin:Hindu 25.6 Obese PN_848 Pune Maharastra 18.52043 73.856744 - 25.6 Obese BN266 Hardoi Uttar Pradesh 27.394647 80.129931 Hindu 25.8 Obese BN051 - Uttar Pradesh 27.570589 80.0981873 Hindu 26.1 Obese

11
PN15_757 Mumbai Maharastra 19.075984 72.877656 Bramhin:Hindu 26.1 Obese PN18_1053 Parola Maharastra 20.87599 75.118393 Bhumiyar:Hindu 26.1 Obese SD191 Adoor Kerala 9.15124 76.730766 Christian 26.12 Obese PN18_1055 Pune Maharastra 18.52043 73.856744 Maratha:Hindu 26.2 Obese PN18_1252 Alibaag Maharastra 18.658173 72.872087 Agari:Hindu 26.3 Obese BN211 Theni Tamil Nadu 10.010366 77.476811 Hindu 26.4 Obese BN254 Thalassery Kerala 11.753288 75.492878 Hindu 26.5 Obese BN260 Wayanad Kerala 11.709446 76.095537 Christian 26.5 Obese BN048 - Kerala 10.850516 76.271083 Hindu 26.6 Obese BN085 - Karnataka 15.317278 75.713888 Hugar:Hindu 26.7 Obese PN18_1047 Pune Maharastra 18.52043 73.856744 Leva_Pail:Hindu 27.1 Obese PN18_1126 - West Bengal 22.586029 88.3587 Hindu 27.3 Obese PN14_721 - Jharkand 23.610181 85.279935 Vaishya:Hindu 27.7 Obese BN053 - Kerala 10.850516 76.271083 Muslim 27.8 Obese BN263 - Himachal Pradesh 31.110375 77.16904 Marwadi:Hindu 28 Obese BN272 - Kerala 10.850516 76.2710833 Christian 28.4 Obese PN15_773 Nashik Maharastra 19.997453 73.789802 Mahar:Hindu 28.7 Obese BN014 Bangalore Karnataka 12.971599 77.594563 Christian 29 Obese BN129 Gulbarga Karnataka 17.329731 76.834296 Hindu 29.4 Obese BN154 Mumbai Maharastra 19.075984 72.877656 Hindu 29.6 Obese BN269 Channapatna Karnataka 12.654186 77.200442 Muslim 29.6 Obese BN100 - Kerala 10.850516 76.2710833 Kshatriya:Hindu 31.7 Obese

12
Supplementary Table S2. Eigenvalue of corresponding eigenvectors with sigma
value of outlier samples
Eigenvector Eigenvalue Number of
outlier samples Sample code Iteration Sigma value 1 1.433229 1 BN100 1 -10.017 2 1.300805 - - - - 3 1.236721 - - - - 4 1.153324 2 SD012, BN059 2, 3 6.628, 6.527 5 1.126309 - - - - 6 1.118402 - - - - 7 1.11018 - - - - 8 1.105451 1 PN14_706 1 -6.123 9 1.101737 - - - - 10 1.099101 - - - -

13
Supplementary Table S3. Details of replication stage (cohort-II) samples.
ID BMI ID BMI ID BMI ID BMI
sample_P6_G02 18 sample_P1_D11 23 sample_P2_A03 18 sample_P2_E04 19
sample_P5_G03 17.6 sample_P1_D12 22 sample_P2_A04 18 sample_P2_E05 19
sample_P7_H09 22.2 sample_P1_E01 16 sample_P2_A05 18 sample_P2_E06 18
sample_P_G03 22.8 sample_P1_E02 16 sample_P2_A06 23 sample_P2_E07 23
sample_P_G07 18 sample_P1_E03 22 sample_P2_A07 23 sample_P2_E08 23
sample_P_H09 29.6 sample_P1_E04 20 sample_P2_A08 22 sample_P2_E09 23
sample_P1_10 22.6 sample_P1_E05 22 sample_P2_A09 24 sample_P2_E10 18
sample_P1_A01 24 sample_P1_E06 24 sample_P2_A10 20 sample_P2_E11 21
sample_P1_A02 24 sample_P1_E07 18 sample_P2_A11 23 sample_P2_E12 19
sample_P1_A03 21 sample_P1_E08 21 sample_P2_A12 21 sample_P2_F01 18
sample_P1_A04 27.3 sample_P1_E09 22 sample_P2_B01 23 sample_P2_F02 15
sample_P1_A07 21.5 sample_P1_E10 22 sample_P2_B02 23 sample_P2_F03 20
sample_P1_A08 21.5 sample_P1_E11 22 sample_P2_B03 19 sample_P2_F04 20
sample_P1_A09 21.2 sample_P1_E12 22 sample_P2_B04 17 sample_P2_F05 21
sample_P1_A10 21.2 sample_P1_F01 24 sample_P2_B05 21 sample_P2_F06 22
sample_P1_B01 19.4 sample_P1_F02 24 sample_P2_B06 20 sample_P2_F07 19
sample_P1_B02 19.2 sample_P1_F03 21 sample_P2_B07 20 sample_P2_F08 19
sample_P1_B03 19.2 sample_P1_F04 21 sample_P2_B08 21 sample_P2_F09 23
sample_P1_B04 21.6 sample_P1_F05 23 sample_P2_B09 27 sample_P2_F10 17
sample_P1_B05 21.6 sample_P1_F06 20 sample_P2_B10 23 sample_P2_F11 25
sample_P1_B06 29 sample_P1_F07 26 sample_P2_B11 18 sample_P2_F12 23
sample_P1_B07 29 sample_P1_F10 27 sample_P2_B12 22 sample_P2_G01 20
sample_P1_B08 22 sample_P1_F11 27 sample_P2_C01 20 sample_P2_G02 19
sample_P1_B09 22 sample_P1_G01 20 sample_P2_C02 20 sample_P2_G03 19
sample_P1_B10 20.8 sample_P1_G02 26 sample_P2_C03 24 sample_P2_G04 19
sample_P1_B11 20.8 sample_P1_G03 26 sample_P2_C04 24 sample_P2_G05 21
sample_P1_B12 24.1 sample_P1_G04 21 sample_P2_C05 24 sample_P2_G06 20
sample_P1_C01 24.1 sample_P1_G05 28 sample_P2_C06 18 sample_P2_G07 21
sample_P1_C02 24.1 sample_P1_G06 21 sample_P2_C07 22 sample_P2_G08 23
sample_P1_C03 24.2 sample_P1_G07 24 sample_P2_C08 23 sample_P2_G09 21
sample_P1_C04 24.2 sample_P1_G08 24 sample_P2_C09 19 sample_P2_G10 21
sample_P1_C05 24.2 sample_P1_G09 19 sample_P2_C10 20 sample_P2_G11 19
sample_P1_C06 19.4 sample_P1_G10 20 sample_P2_C11 22 sample_P2_G12 19
sample_P1_C07 17.6 sample_P1_G11 22 sample_P2_C12 22 sample_P2_H01 19
sample_P1_C08 17 sample_P1_G12 20 sample_P2_D01 20 sample_P2_H02 20
sample_P1_C08 17.6 sample_P1_H01 20 sample_P2_D02 21 sample_P2_H03 21
sample_P1_C09 17.6 sample_P1_H02 18 sample_P2_D03 32 sample_P2_H04 21
sample_P1_C10 27.6 sample_P1_H03 18 sample_P2_D04 21 sample_P2_H05 22
sample_P1_C11 20.3 sample_P1_H04 21 sample_P2_D05 25 sample_P2_H06 23
sample_P1_D01 18.8 sample_P1_H06 24 sample_P2_D06 22 sample_P2_H07 18
sample_P1_D02 23.1 sample_P1_H07 23 sample_P2_D07 20 sample_P2_H08 16
sample_P1_D03 23.1 sample_P1_H08 23 sample_P2_D08 20 sample_P2_H09 30
sample_P1_D04 22.8 sample_P1_H09 20 sample_P2_D09 25 sample_P2_H10 23
sample_P1_D05 22.8 sample_P1_H10 23 sample_P2_D10 18 sample_P2_H11 20
sample_P1_D06 19.8 sample_P1_H11 23 sample_P2_D11 19 sample_P2_H12 20

14
sample_P1_D07 19.8 sample_P1_H12 18 sample_P2_D12 19 sample_P3_A01 24
sample_P1_D08 21.6 sample_P2_10 23 sample_P2_E01 20 sample_P3_A02 22
sample_P1_D09 18.7 sample_P2_A01 18 sample_P2_E02 18 sample_P3_A04 16
sample_P1_D10 18.1 sample_P2_A02 18 sample_P2_E03 20 sample_P3_A07 17
ID BMI ID BMI ID BMI ID BMI
sample_P3_A08 18.1 sample_P3_F01 16.1 sample_P4_B06 26.8 sample_P4_F07 23.7
sample_P3_A09 18.4 sample_P3_F02 23.2 sample_P4_B07 18.9 sample_P4_F08 17.9
sample_P3_A10 20.8 sample_P3_F03 20.1 sample_P4_B08 20.2 sample_P4_F10 16.4
sample_P3_A11 21.5 sample_P3_F04 20.1 sample_P4_B09 26.4 sample_P4_F11 22.2
sample_P3_B01 37.3 sample_P3_F05 26 sample_P4_B10 18.1 sample_P4_F12 21.3
sample_P3_B02 19.8 sample_P3_F06 24.6 sample_P4_B11 17.3 sample_P4_G01 17.3
sample_P3_B03 20 sample_P3_F07 18.1 sample_P4_B12 19.8 sample_P4_G02 20
sample_P3_B04 20 sample_P3_F09 18.6 sample_P4_C01 23.3 sample_P4_G03 19.1
sample_P3_B05 23.4 sample_P3_F10 23.8 sample_P4_C02 17.7 sample_P4_G04 19.4
sample_P3_B06 23.4 sample_P3_F11 26.8 sample_P4_C03 21.7 sample_P4_G05 25.8
sample_P3_B07 19.4 sample_P3_F12 18.3 sample_P4_C04 26.2 sample_P4_G06 23.7
sample_P3_B08 22 sample_P3_G01 22.3 sample_P4_C05 17.6 sample_P4_G07 24
sample_P3_B09 17.6 sample_P3_G02 19.5 sample_P4_C06 22.5 sample_P4_G08 24.5
sample_P3_B10 20.5 sample_P3_G03 26.5 sample_P4_C07 25.8 sample_P4_G09 25.4
sample_P3_B11 23.4 sample_P3_G04 26.5 sample_P4_C08 20.5 sample_P4_G10 17.4
sample_P3_B12 23.4 sample_P3_G05 20.2 sample_P4_C09 20.8 sample_P4_G11 26.1
sample_P3_C01 20.1 sample_P3_G06 20.2 sample_P4_C10 17.5 sample_P4_G12 18.5
sample_P3_C02 23.9 sample_P3_G07 21.7 sample_P4_C11 27.3 sample_P4_H01 18.5
sample_P3_C03 21.3 sample_P3_G08 24.1 sample_P4_C12 28.4 sample_P4_H02 23.1
sample_P3_C04 24.9 sample_P3_G09 27.3 sample_P4_D01 17.6 sample_P4_H03 19.5
sample_P3_C05 19.7 sample_P3_G10 20.9 sample_P4_D02 24 sample_P4_H04 18.8
sample_P3_C06 19.7 sample_P3_G11 21.3 sample_P4_D03 22.7 sample_P4_H05 17.6
sample_P3_C07 22 sample_P3_G12 24.2 sample_P4_D04 20.3 sample_P4_H06 19.4
sample_P3_C08 22 sample_P3_H01 24.4 sample_P4_D05 16.7 sample_P4_H07 21.2
sample_P3_C09 25.6 sample_P3_H02 18.4 sample_P4_D06 20.7 sample_P4_H08 18.1
sample_P3_C10 25.6 sample_P3_H03 22.8 sample_P4_D07 18.7 sample_P4_H09 18.4
sample_P3_C11 21.5 sample_P3_H04 19.1 sample_P4_D08 18.6 sample_P4_H10 23.7
sample_P3_D03 21.5 sample_P3_H05 16 sample_P4_D09 18.4 sample_P4_H11 15.2
sample_P3_D04 21.5 sample_P3_H06 26.9 sample_P4_D10 19 sample_P4_H12 21.5
sample_P3_D05 22.9 sample_P3_H07 25.6 sample_P4_D11 20.9 sample_P5_A01 21.5
sample_P3_D06 22.9 sample_P3_H08 16.9 sample_P4_D12 18.4 sample_P5_A02 22.2
sample_P3_D07 17.6 sample_P3_H09 19.9 sample_P4_E01 20.7 sample_P5_A03 18.5
sample_P3_D08 24.8 sample_P3_H10 18.2 sample_P4_E02 21.4 sample_P5_A04 23.7
sample_P3_D09 24.8 sample_P3_H12 24.8 sample_P4_E03 15.7 sample_P5_A05 21.8
sample_P3_D10 17.5 sample_P4_A01 19.6 sample_P4_E04 18.3 sample_P5_A06 25.2
sample_P3_D11 20.6 sample_P4_A02 18.6 sample_P4_E05 16.6 sample_P5_A07 27.7
sample_P3_D12 20.4 sample_P4_A03 17.4 sample_P4_E06 28.7 sample_P5_A08 21.9
sample_P3_E01 23.9 sample_P4_A04 25.5 sample_P4_E07 23.1 sample_P5_A09 18.5
sample_P3_E02 18.9 sample_P4_A05 22.5 sample_P4_E08 21 sample_P5_A10 16.1
sample_P3_E03 19.5 sample_P4_A06 18.6 sample_P4_E09 22.7 sample_P5_A11 20.2
sample_P3_E04 14.2 sample_P4_A07 20.8 sample_P4_E10 19.1 sample_P5_A12 21.3
sample_P3_E05 26.4 sample_P4_A08 20.1 sample_P4_E11 18 sample_P5_B01 20.8
sample_P3_E06 22.9 sample_P4_A09 15.6 sample_P4_E12 26.6 sample_P5_B02 23.5

15
sample_P3_E07 26 sample_P4_A10 18.6 sample_P4_F01 23.3 sample_P5_B03 17.7
sample_P3_E08 22.9 sample_P4_A11 18.3 sample_P4_F02 25.6 sample_P5_B04 18.4
sample_P3_E09 24.9 sample_P4_A12 16.9 sample_P4_F03 17.3 sample_P5_B05 23.9
sample_P3_E10 16.4 sample_P4_B02 18.3 sample_P4_F04 15.4 sample_P5_B06 21.1
sample_P3_E11 24.9 sample_P4_B03 17.3 sample_P4_F05 16.9 sample_P5_B07 22.6
sample_P3_E12 25.8 sample_P4_B04 18.4 sample_P4_F06 16.6 sample_P5_B08 20.4
ID BMI ID BMI ID BMI ID BMI
sample_P5_B09 19.6 sample_P5_F10 18.1 sample_P6_B11 23.3 sample_P6_G01 19.1
sample_P5_B10 19 sample_P5_F11 18.3 sample_P6_B12 20.2 sample_P6_G02 18
sample_P5_B11 25 sample_P5_F12 21.8 sample_P6_C01 18.9 sample_P6_G03 22.8
sample_P5_B12 19.8 sample_P5_F4 18.4 sample_P6_C02 22.3 sample_P6_G04 16.4
sample_P5_C01 19 sample_P5_G01 22.6 sample_P6_C03 25.2 sample_P6_G05 17.3
sample_P5_C02 22.7 sample_P5_G02 23.7 sample_P6_C04 21.4 sample_P6_G06 21.6
sample_P5_C03 16.4 sample_P5_G03 17.6 sample_P6_C06 23.3 sample_P6_G07 23.5
sample_P5_C04 17.7 sample_P5_G04 20.7 sample_P6_C07 23.3 sample_P6_G08 18.7
sample_P5_C05 22.2 sample_P5_G05 18.9 sample_P6_C08 25.1 sample_P6_G09 26.9
sample_P5_C06 18 sample_P5_G06 21.6 sample_P6_C09 25.1 sample_P6_G10 19.5
sample_P5_C07 18.2 sample_P5_G07 26.2 sample_P6_C10 20.7 sample_P6_G11 19.5
sample_P5_C08 24.1 sample_P5_G08 26.1 sample_P6_C11 20.7 sample_P6_G12 18.3
sample_P5_C09 24.1 sample_P5_G09 18.1 sample_P6_C12 20.7 sample_P6_H 18
sample_P5_C10 21.8 sample_P5_G10 19 sample_P6_D01 16.3 sample_P6_H01 23.4
sample_P5_C11 20.8 sample_P5_G11 34.5 sample_P6_D02 16.3 sample_P6_H02 22.8
sample_P5_C12 20.6 sample_P5_G12 24.2 sample_P6_D03 16.3 sample_P6_H03 17
sample_P5_D01 21 sample_P5_H01 28.3 sample_P6_D04 19.7 sample_P6_H04 23
sample_P5_D02 27.8 sample_P5_H02 25.5 sample_P6_D05 19.7 sample_P6_H05 26.2
sample_P5_D03 19.1 sample_P5_H04 18.2 sample_P6_D06 24 sample_P6_H06 17.2
sample_P5_D04 17.4 sample_P5_H05 18 sample_P6_D07 24 sample_P6_H07 21
sample_P5_D05 24.5 sample_P5_H06 18 sample_P6_D08 19 sample_P6_H08 18
sample_P5_D06 21.2 sample_P5_H07 19.2 sample_P6_D09 19 sample_P6_H09 17.2
sample_P5_D07 20.8 sample_P5_H08 18.2 sample_P6_D10 23.5 sample_P6_H10 20.7
sample_P5_D08 21.5 sample_P5_H09 17.3 sample_P6_D11 23.5 sample_P6_H11 20.7
sample_P5_D09 19.5 sample_P5_H10 24.6 sample_P6_D12 19.8 sample_P6_H12 20.7
sample_P5_D10 23.7 sample_P5_H11 28 sample_P6_E01 19.8 sample_P7_A03 22.8
sample_P5_D11 18.4 sample_P5_H12 24.2 sample_P6_E02 23.3 sample_P7_A04 18.7
sample_P5_D12 22.5 sample_P6_A01 18.1 sample_P6_E03 23.3 sample_P7_A05 20.4
sample_P5_E01 16.6 sample_P6_A02 21.3 sample_P6_E04 23.9 sample_P7_A06 26.6
sample_P5_E02 16.6 sample_P6_A03 16.5 sample_P6_E05 27.3 sample_P7_A07 26.3
sample_P5_E03 30.2 sample_P6_A04 16.5 sample_P6_E06 17.9 sample_P7_A08 23.3
sample_P5_E04 25.3 sample_P6_A05 19.1 sample_P6_E07 22.7 sample_P7_A09 26.8
sample_P5_E05 24.3 sample_P6_A06 15.4 sample_P6_E08 27.3 sample_P7_A10 26.8
sample_P5_E06 24.2 sample_P6_A07 15.4 sample_P6_E09 17.9 sample_P7_A11 26.8
sample_P5_E07 19.8 sample_P6_A08 22 sample_P6_E10 22 sample_P7_A12 17.6
sample_P5_E08 27.9 sample_P6_A09 17.9 sample_P6_E11 17.9 sample_P7_B01 17.6
sample_P5_E09 18.4 sample_P6_A10 20.6 sample_P6_E12 22.8 sample_P7_B02 17.6
sample_P5_E10 18.4 sample_P6_A11 20.6 sample_P6_F01 22.8 sample_P7_B03 21.3
sample_P5_E11 26.4 sample_P6_A12 18.9 sample_P6_F02 18.5 sample_P7_B05 25.2

16
sample_P5_E12 17 sample_P6_B01 27.1 sample_P6_F03 19 sample_P7_B06 21.38
sample_P5_F01 21.6 sample_P6_B02 23.1 sample_P6_F04 19 sample_P7_B07 23.5
sample_P5_F02 18 sample_P6_B03 17.6 sample_P6_F05 23.5 sample_P7_B08 25.14
sample_P5_F03 26.1 sample_P6_B04 22.3 sample_P6_F06 23.5 sample_P7_B09 26.17
sample_P5_F04 18.4 sample_P6_B05 26.1 sample_P6_F07 21.7 sample_P7_B10 24.22
sample_P5_F05 24.6 sample_P6_B06 18.8 sample_P6_F08 21.7 sample_P7_B11 26.79
sample_P5_F06 24.6 sample_P6_B07 26.2 sample_P6_F09 27.4 sample_P7_B12 20.8
sample_P5_F07 23.9 sample_P6_B08 28.4 sample_P6_F10 27.4 sample_P7_C01 35.05
sample_P5_F08 21.8 sample_P6_B09 20.2 sample_P6_F11 24.3 sample_P7_C02 27.21
sample_P5_F09 23.7 sample_P6_B10 21.1 sample_P6_F12 23.7 sample_P7_C03 23.87
ID BMI ID BMI
sample_P7_C04 21.9 sample_P7_G08 20.81
sample_P7_C05 24.52 sample_P7_G09 19.6
sample_P7_C06 22.72 sample_P7_G10 18.37
sample_P7_C07 20.98 sample_P7_G11 17.48
sample_P7_C08 15.49 sample_P7_G12 19.85
sample_P7_C09 23.93 sample_P7_H01 19.84
sample_P7_C10 24.16 sample_P7_H02 20.76
sample_P7_C11 20.07 sample_P7_H02 76
sample_P7_C12 16.33 sample_P7_H03 20.31
sample_P7_D01 19.08 sample_P7_H04 24.22
sample_P7_D02 20.13 sample_P7_H05 19.46
sample_P7_D04 24.22 sample_P7_H06 19.71
sample_P7_D05 14.01 sample_P7_H07 24.81
sample_P7_D06 23.1 sample_P7_H08 19.46
sample_P7_D07 22.73 sample_P7_H09 22.22
sample_P7_D09 16.33 sample_P7_H10 23.78
sample_P7_D10 25.39 sample_P7_H11 26.12
sample_P7_D11 23.93 sample_P7_H12 20.61
sample_P7_D12 17.99 sample_P7_E01 19.85 sample_P7_E02 22.53 sample_P7_E03 21.67 sample_P7_E04 22.38 sample_P7_E05 23.38 sample_P7_E06 21.35 sample_P7_E07 22.09 sample_P7_E08 20.82 sample_P7_E09 22.32 sample_P7_E10 18.16 sample_P7_E11 26.12 sample_P7_E12 19.83 sample_P7_F01 21.63 sample_P7_F03 24.48 sample_P7_F04 25.39 sample_P7_F05 25.2 sample_P7_F06 23.72

17
sample_P7_F07 23.72 sample_P7_F08 23.93 sample_P7_F09 22.06 sample_P7_F10 22.53 sample_P7_F11 22.72 sample_P7_F12 22.72 sample_P7_G01 19.08 sample_P7_G02 23.03 sample_P7_G03 23.03 sample_P7_G04 20.2 sample_P7_G05 14.01 sample_P7_G06 19.88 sample_P7_G07 17.99

18
Supplementary Table S4. Details of SNPs having extreme p-value (p≤ 3.75×10-5
) in Discovery stage (cohort-I) and 4
SNPs used in replication study.
Stage Chr RsID Position (hg19) Gene β S.E. R2 T P-value HWE P-value N
Discovery stage 21 rs2406338 19929628 - - 1.665 0.343 0.1044 4.853 2.43E-06 0.01965 204 Discovery stage 13 rs9521445 110285534 - - -1.459 0.3065 0.1009 -4.761 3.67E-06 0.7598 204 Discovery stage 13 rs7989848 110283468 - - -1.426 0.3034 0.09861 -4.701 4.79E-06 0.6502 204 Discovery stage 7 rs12699560 13835262 - - 1.865 0.3997 0.09731 4.666 5.57E-06 0.1189 204 Discovery stage 21 rs2827881 24505930 - - 1.469 0.3156 0.09689 4.655 5.85E-06 0.2524 204 Discovery stage 21 rs1304747 24585237 - - -1.385 0.2994 0.09577 -4.625 6.67E-06 0.2894 204 Discovery stage 2 rs12987839 24688055 - - 1.323 0.3019 0.08679 4.381 1.89E-05 1 204 Discovery stage 16 rs11645605 79047468 WWOX Intronic -1.708 0.3921 0.08583 -4.355 2.11E-05 0.1366 204 Discovery stage 7 rs2051930 13836003 - - 1.751 0.4023 0.08572 4.352 2.14E-05 0.1189 204 Discovery stage 13 rs9551868 30783104 KATNAL1 Intronic -1.559 0.3587 0.08547 -4.345 2.21E-05 0.2798 204 Discovery stage 21 rs9680422 24590183 - - -1.322 0.3047 0.08529 -4.34 2.25E-05 0.3593 204 Discovery stage 1 rs2281524 93602194 MTF2 Intronic -1.48 0.3414 0.08517 -4.337 2.28E-05 0.2346 204 Discovery stage 7 rs1526538 11774241 THSD7A Intronic -1.406 0.3247 0.08495 -4.33 2.34E-05 0.04954 204 Discovery stage 19 rs17796053 16371818 - - 1.707 0.3966 0.08402 4.305 2.61E-05 0.05072 204 Discovery stage 9 rs10821443 93192455 - - 1.36 0.317 0.0835 4.29 2.77E-05 0.2546 204 Discovery stage 17 rs236594 68207627 - - 1.519 0.3548 0.08316 4.28 2.88E-05 0.7073 204 Discovery stage 1 rs2815429 93590668 MTF2 Intronic -1.549 0.3622 0.083 -4.276 2.94E-05 0.8364 204 Discovery stage 4 rs7681875 41556422 LIMCH1 Intronic -1.294 0.3049 0.08192 -4.246 3.32E-05 1 204 Discovery stage 19 rs11671078 16373071 - - 1.328 0.313 0.0818 4.242 3.37E-05 1 204 Discovery stage 13 rs2031998 30783452 KATNAL1 Intronic -1.526 0.3599 0.08168 -4.239 3.42E-05 0.2726 204 Discovery stage 1 rs2632300 98616100 - - 1.264 0.2999 0.08088 4.216 3.75E-05 1 204 Discovery stage 4 rs7682470 41556709 LIMCH1 Intronic -1.291 0.3061 0.08087 -4.216 3.75E-05 0.8869 204 Replication stage 13 rs9551868 30783104 KATNAL1 Intronic -0.4602 0.3932 0.00467 -1.17 0.2428 4.058E-05 214* Replication stage 1 rs2281524 93602194 MTF2 Intronic -0.3875 0.2542 0.004857 -1.524 0.1281 0.887 478* Replication stage 7 rs1526538 11774241 THSD7A Intronic -0.8791 0.1957 0.04176 -4.492 8.92E-06 0.1376 465* Replication stage 4 rs7681875 41556422 LIMCH1 Intronic -0.5889 0.2282 0.01542 -2.58 0.01021 1 427*
* Out of 654, few samples are removed due to bad quality of sequences (noise in the electropherograms)

19
Supplementary Table S5.Sequence of primers and their PCR condition.
Primer Gene Forward Sequence Reverse Sequence
Co
nc. (p
m/µ
l)
Temperature: °C (time: minute);
total 35 cycles
Init
ial
den
atu
rati
on
Den
atu
rati
on
An
nea
lin
g
Ex
ten
sio
n
Fin
al
Ex
ten
sio
n
rs9551868 KATNAL1 TGCCTTCAGTTGCCTACAAATACTCCA AAGAAACATCCTGGGGGTTCAGCA 0.4 95 (5) 95 (2) 60 (0.5) 72 (2) 72(7) rs7681875 LIMCH1 GGCTGAATTGGTGGCTCTGCT TGGTTCCCCTGCAGTTTTTACTGGT 0.4 95 (5) 95 (2) 60 (0.5) 72 (2) 72(7) rs1526538 THSD7A AGGCTTTTCATCCTCTACACTCCAGA TGGTGAAGCACTGTGTTATGCCTTGT 0.5 95 (5) 95 (2) 60 (0.5) 72 (2) 72(7) rs2281524 MTF2 GCTCCACAAAGTTCAGCTCCAACA TCGCCACATGCTATTCTACCTGC 0.5 95 (5) 95 (2) 60 (0.5) 72 (2) 72(7)

20
Supplementary Table S6. Mean and standard deviation of BMI stratified
according to genotype.
SNP
Gen
oty
pe
Co
un
ts
Fre
qu
ency
BM
I-m
ean
BM
I-st
and
ard
dev
iati
on
Co
un
ts
Fre
qu
ency
BM
I-m
ean
BM
I-st
and
ard
dev
iati
on
Co
un
ts
Fre
qu
ency
BM
I-m
ean
BM
I-st
and
ard
dev
iati
on
Discovery stage Replication stage Combined
rs9551868 AA 11 0.05392 18.77 2.905
Not in Hardy-Weinberg equilibrium AT 61 0.299 20.85 3.012 TT 132 0.6471 22.24 3.051
rs2281524 CC 14 0.06863 19.02 2.21 21 0.04393 20.81 2.511 35 0.05132 20.09 2.524 CT 66 0.3235 20.98 3.268 155 0.3243 21.22 3.388 221 0.324 21.15 3.347 TT 124 0.6078 22.28 2.979 302 0.6318 21.6 3.137 426 0.6246 21.8 3.103
rs1526538 GG 43 0.2108 20.25 2.788 121 0.2602 20.49 2.977 164 0.2451 20.43 2.922 GA 117 0.5735 21.6 3.053 216 0.4645 21.1 3.019 333 0.4978 21.28 3.036 AA 44 0.2157 23.06 3.216 128 0.2753 22.24 3.3 172 0.2571 22.45 3.289
rs7681875 GA 36 0.1765 20.43 2.634 70 0.1639 20.65 3.247 106 0.168 20.58 3.042 GA 100 0.4902 21.22 2.939 205 0.4801 21.2 3.065 305 0.4834 21.21 3.02 AA 68 0.3333 22.88 3.349 152 0.356 21.82 3.568 220 0.3487 22.15 3.528

21
Supplementary Table S7. Power of association of THSD7A gene and
previously reported SNPs with BMI in the present study.
Ch
r
rsID
-Cau
sal
var
ian
t
All
ele
All
ele
freq
uen
cy
Nu
mb
er o
f
sub
ject
s
Gen
e
Ref
.
Bet
a v
alu
e
Po
wer
(p
-val
ue
=
0.0
5)
19 rs11084753-G G 0.5588 204 KCTD15 - RPS4XP20 2 0.06 0.0504 1 rs1514175-A A 0.5196 204 TNNI3K;FPGT-TNNI3K 1 0.07 0.0506 1 rs1555543-C C 0.5613 204 EEF1A1P11 - NDUFS5P2 1 0.06 0.0504 18 rs17782313-C C 0.3529 204 RPS3AP49 - MC4R 2 0.2 0.0543 1 rs2815752-A A 0.6127 204 RPL31P12 - KRT8P21 2 0.1 0.0511 1 rs2815752-A A 0.6127 204 RPL31P12 - KRT8P21 1 0.13 0.0519 11 rs3817334-T T 0.348 204 MTCH2 1 0.06 0.0504 1 rs543874-G G 0.1667 204 BRINP2 - SEC16B 1 0.22 0.0531 18 rs571312-A A 0.3113 204 RPS3AP49 - MC4R 1 0.23 0.0553 2 rs6548238-C C 0.8652 204 FAM150B - TMEM18 2 0.26 0.0537 2 rs713586-C C 0.4804 204 ADCY3 - DNAJC27 1 0.14 0.0523 12 rs7138803-A A 0.375 204 BCDIN3D - RPL35AP28 1 0.12 0.0516 2 rs939583-C C 0.2034 204 FAM150B - TMEM18 3 0.07 0.0504 6 rs987237-G G 0.2059 204 TFAP2B 1 0.13 0.0513 16 rs9939609-A A 0.6912 204 FTO 4 0.36 0.063 16 rs9939609-A A 0.6912 204 FTO 2 0.33 0.0609

22
References
1. Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al.
Association analyses of 249,796 individuals reveal 18 new loci associated with body
mass index. Nat Genet 2010; 42: 937–948.
2. Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM et al. Six new loci
associated with body mass index highlight a neuronal influence on body weight
regulation. Nat Genet 2009; 41:25–34.
3. Pei YF, Zhang L, Liu Y, Li J, Shen H, Liu YZ et al. Meta-analysis of genome-wide
association data identifies no vel susceptibility loci for obesity. Hum Mol Genet 2014;
23:820–830.
4. Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM et al.
A common variant in the FTO gene is associated with body mass index and
predisposes to childhood and adult obesity. Science 2007; 316: 889–894.