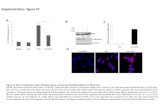
Supplementary Figure A2. Multi-image registration scheme ...€¦ · Supplementary Figure D1....
Transcript of Supplementary Figure A2. Multi-image registration scheme ...€¦ · Supplementary Figure D1....

1
Supplementary material for van de Schoot AJAJ, et al. Quantification of delineation errors of the gross tumor volume on magnetic resonance imaging in uterine cervical cancer using pathology data and deformation cor-rection, Acta Oncol 2015;54:224–31.
Details on weight factor optimization
To correlate MRi and pathology data for our pur-pose, we need to find the spatial relation between the pathology photo (IPAT) and its corresponding MR image (IMRI) described by a transformation (T). This transformation is determined by a similarity optimi-zation using a cost function (C) to describe this similarity and in general performed for only one moving image to align one fixed image. however, the process of image registration in this study was per-formed using multiple image pairs. given multiple imaging pairs, all image pairs were simultaneously
Supplementary Figure A1. An example of a 2D digital photo of the macroscopic intersection (a) and the corresponding 2D sagittal MR image (c) and with delineated structures [cervix-uterus (blue), uterine cavity (green), gTv (red)] (b, d). The delineated structures were segmented from the background on the photo (e–g) and from surrounding tissue on MRi (h–j) and transformed to binary images (k–p).
Supplementary Figure A2. Multi-image registration scheme between segmentations of MRi [cervix-uterus (a), uterine cavity (b)] and the pathology photo [cervix-uterus (c), uterine cavity (d)]. The pathology segmentations were registered to the MRi segmentations and pathology images were deformed (e–f) using the obtained transformation T.
registered using a single type of cost function by combining cost function values of each image pair based on pre-defined weight factors (wi) during the optimization process (Equation 1).
in this study, the second and third (i.e. affine and deformable) registration step of the three-step multi-image registration strategy were performed using multiple imaging sets (i.e. one image pair of the cer-vix-uterus and one image pair of the uterine cavity). To obtain the most accurate correlation between both imaging sets, the optimal contribution of each image pair to the final transformation was

2
Supplementary Figure B2. Median DSC (a) and SDE (b) of the cervix-uterus structure (red solid line) and the uterine cavity structure (blue dashed line) for all patients after three-step multi-image registration as function of the used contributions of each image pair (wCU - wUCa). The error bars indicate the 25%–75% range. Based on all 16 patients, an equal contribution of both image pairs (wCU 0.5; wUCa 0.5) was determined to be optimal.
Supplementary Figure B1. Average DSC (a) and average SDE (b) of the cervix-uterus structure and the uterine cavity structure after three-step multi-image registration as function of the used contributions of each image pair (wCU - wUCa) for three different patients. The optimal contribution of both image pairs (wCU - wUCa) to the final registration was indicated for the three presented patients.
determined. Therefore, three-step multi-image regis-trations were performed multiple times with the con-tribution of the cervix-uterus image pair (wCU) and the uterine cavity image pair (wUCa) varied between no contribution (wi 0.0) and complete contribution (wi 1.0).
After three-step multi-image registrations using different contributions of each image pair, the cor-relation between corresponding structures in terms of DSC and SDE was evaluated. For each patient, both the DSC and the SDE of the cervix-uterus structure and the uterine cavity structure was calcu-
lated and average combinations of both structures were plotted (Supplementary Figure B1). given these results the optimal contribution of each image pair, indicated by the maximum DSC and minimum SDE, was determined and used as patient-specific weight factor. Finally, the DSC and the SDE of the cervix-uterus structure and the uterine cavity struc-ture based on the entire population (N 16) were calculated (Supplementary Figure B2). The popula-tion-based optimal contribution of each image pair to the final transformation (wCU 0.5; wUCa 0.5) can be used for ensuing patients.

3
Supplementary Figure D1. Box-and-whisker plots of the DSC and the SDE between MRi and pathology data (REg2) after each step (rigid registration, affine registration and deformable registration) in the three-step multi-image registration strategy for the cervix-uterus and the uterine cavity structure. The box represents the upper and lower quartiles (iQR) and the band (red) inside the box the median value. The whisker represents the highest (lowest) value within 1.5 iQR of the upper (lower) quartile. Dots above or below the whiskers are considered outliers. horizontal lines indicate statistical significant difference (p 0.01).
Supplementary Figure C1. Schematic (left) and MRi-based (right) representation of gTvMRi expansions. The delineated gTv (solid red line) is expanded uniformly only within the cervix-uterus structure (blue) using increasing margins (dashed blue lines).

4
Supplementary Table E1. Mean (SD) absolute pixel displacement (mm) inside different structures caused by the deformable multi-image registration step after REg1 and REg2.
Patient #
Cervix-Uterus Uterine Cavity gTv
REg1 REg2 REg1 REg2 REg1 REg2
1 6.5 (2.5) 7.3 (3.0) 8.8 (1.6) 10.3 (2.3) 2.6 (1.2) 3.2 (1.4)2 8.3 (3.4) 8.0 (3.7) 10.9 (4.3) 10.6 (4.8) 9.6 (2.0) 9.0 (1.7)3 3.8 (2.1) 5.0 (2.8) 3.1 (1.2) 5.2 (3.0) 3.3 (1.9) 2.9 (1.8)4 2.1 (1.0) 3.8 (2.0) 2.5 (0.4) 9.4 (0.9) 2.6 (0.8) 3.8 (1.1)5 4.1 (2.0) 8.2 (3.9) 7.5 (0.1) 9.6 (0.3) 4.2 (2.1) 11.2 (1.4)6 6.2 (2.7) 5.8 (2.8) 10.5 (0.7) 10.1 (0.5) 5.4 (1.3) 4.0 (1.8)7 3.2 (1.9) 7.8 (3.3) 3.9 (1.9) 9.7 (3.9) 1.1 (0.3) 8.3 (0.7)8 4.9 (3.2) 5.2 (2.9) 4.7 (2.4) 3.8 (2.3) 3.7 (1.8) 4.6 (2.1)9 5.2 (2.7) 7.8 (2.3) 3.6 (1.8) 8.7 (1.7) 6.4 (1.5) 7.7 (2.6)
10 7.0 (2.5) 8.0 (4.5) 7.8 (1.9) 5.7 (4.0) 5.1 (0.8) 10.5 (4.0)11 3.6 (2.8) 5.2 (4.0) 2.9 (2.6) 4.9 (3.0) 5.7 (1.0) 7.6 (0.8)12 8.2 (2.9) 8.2 (4.1) 10.0 (2.3) 11.2 (4.5) 9.2 (2.2) 7.7 (3.0)13 2.7 (1.2) 3.2 (1.6) 3.1 (1.0) 3.0 (1.2) 2.3 (0.8) 3.4 (1.6)14 3.7 (3.8) 4.0 (4.0) 1.4 (0.5) 1.3 (0.6) 5.4 (3.8) 6.2 (3.5)15 4.3 (1.9) 4.7 (2.0) 4.2 (1.5) 4.2 (1.3) 2.9 (1.5) 3.8 (2.1)16 3.8 (1.9) 3.9 (2.1) 3.4 (1.8) 3.7 (2.6) 5.3 (1.5) 6.3 (1.6)Overall 4.8 (2.4) 6.0 (3.1) 5.5 (1.6) 7.0 (2.3) 4.7 (1.5) 6.3 (1.9)
gTv, gross tumor volume; mm, millimeter; SD, standard deviation.
Supplementary Figure F1. Median gTv coverage after three-step multi-image registration as a function of applied margin to the gTvMRi within the cervix-uterus structure. The red line (squares) shows the coverage of the gTvMRi-2 based on the transformation obtained after REg1 and the blue line (circles) shows the coverage of the gTvMRi-1 based on the transformation obtained after REg2. The black dashed line represents the average median gTv coverage of both observations after only rigid registration and the gray region indicates the 95% confidence interval of both observations after three-step multi-image registration.