Sequence formats and databases in...
Transcript of Sequence formats and databases in...
Sequence formats and databases in bioinformatics
• Definitions/Basics
• Sequence formats
• Databases in Biology
Dinesh GuptaStructural and Computational Biology [email protected]
What is Bioinformatics?
•Bioinformatics is the use of computers to solve biological and
biomedical problems.
•Bioinformatics is the application of information technology to mine,
visualize, analyze, integrate, and manage biological and genetic
information, which can then be applied in, among other things,
accelerating drug discovery and development.
•Application of tools of computation and analysis to the capture and
interpretation of biological data.
•Biological Data management and analysis.
•NIH definition of Bioinformatics (http://www.bisti.nih.gov/CompuBioDef.pdf)
Research, development, or application of computational tools and
approaches for expanding the use of biological, medical,
behavioral or health data, including those to acquire, store,
organize, archive, analyze, or visualize such data.
Use of Bioinformatics
• DNA analysis
– Genome sequencing
• Sequence assembly
• Sequence/gene annotations
• Genefinding/Sequence translation tools
• Sequence Similarity searching (eg. BLAST,
ClustalW)
• Comparison between genomes
• Evolution of sequences (Phylogenetic analysis)
• Gene expression
• Protein analysis
– Structure
• X-ray crystallography
• Homology based models
• Drug designing
– Sequence
• Sequence similarity
• Protein family assignments
• Conserved motifs
• Proteomics data analysis
• Protein Evolution
Use of Bioinformatics (..contd.)
• Other uses:
– Drug designing
– Vaccine development
– Dairy technology
– Forensics
– Crop improvement
– Designing enzymes for detergents
– Genetic counseling
Uses of Bioinformatics (..contd.)
Bioinformatics: Integration of several fields
Bioinformatics
Computer
Science
Mathematics
Statistics
Chemistry
Physics
Biological
Science
Recent events making bioinformatics more important
• Exponential expansion of biological information
• Expansion of multiple types of information
• Cheaper high throughput technologies
• Improvement in computation power
• Lack of standards/quality
• Need for micro and macro analysis
• Need for better algorithms
Vast Growth in (Structural)
Data...
but number of Fundementally
New (Fold) Parts Not
Increasing that Fast
New Submissions
New Folds
Total in Databank
Bioinformatics Analysis?
It is like any other lab analysis!
• You need to know your data/input sources
• You need to understand your methods and their assumptions
• You need a plan to get from point A to point B
• You need to understand your equipment
• You need to be critical and understand potential sources of error
• You need to interpret your results
• Your results need to be reproducible
• Your results should be testable
References, but not limited to:-
• http://www.ncbi.nlm.nih.gov/About/primer/bioinformatics.html
• http://icgeb.res.in/whotdr
• http://en.wikipedia.org/wiki/Bioinformatics
• Baxevanis & Ouellette 2001. Bioinformatics: A Practical Guide to the
Analysis of Genes and Proteins 2nd Edition. John Wiley Publishing.
• Gibas & Jambeck 2001. Developing Bioinformatics Computer Skills.
O’Reilly.
• Bioinformatics: Genome Sequence Analysis Mount 2001
• Bioinformatics For Dummies – Claverie & Notredame 2003
• Introduction to Bioinformatics – Lesk 2002
Sequence formats: Basics
• Why different formats?
– Type of information
– Software requirements
– Database requirements
Main file formats used in Bioinformatics
•ASN.1
•EMBL, Swiss Prot
•FASTA
•GCG
•GenBank/GenPept
•PHYLIP
•PIR
ASN 1: Abstract Syntax Notation 1used by NCBI
Seq-entry ::= set {
class phy-set ,
descr {
pub {
pub {
article {
title {
name "Cross-species infection of blood parasites between resident
and migratory songbirds in Africa" } ,
authors {
names
std {
{
name
name {
last "Waldenstroem" ,
first "Jonas" ,
initials "J." } } ,
{
name
name {
last "Bensch" ,
first "Staffan" ,
initials "S." } } ,
{
name
name {
last "Kiboi" ,
first "Sam" ,
initials "S." } } ,
{
name
name {
last "Hasselquist" ,
first "Dennis" ,
initials "D." } } ,
{
name
name {
last "Ottosson" ,
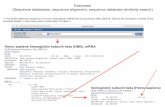
• The first line of each sequence entry is the ID definition line which contains entry name,
dataclass, molecule, division and sequence length.
• XX line contains no data, just a separator
• The AC line lists the accession number.
• DE line gives description about the sequence
• FT precise annotation for the sequence
• Sequence information SQ in the first two spaces.
• The sequence information begins on the fifth line of the sequence entry.
• The last line of each sequence entry in the file is a terminator line which has the two
characters // in the first two spaces.
ID AA03518 standard; DNA; FUN; 237 BP. XX AC U03518;
XX
AC U03518;
XX
DE Aspergillus awamori internal transcribed spacer 1 (ITS1) and 18S
DE rRNA and 5.8S rRNA genes, partial sequence.
DE rRNA and 5.8S rRNA genes, partial sequence.
RX MEDLINE; 94303342.
RX PUBMED; 8030378.
XX
FT rRNA <1..20
FT /product="18S ribosomal RNA"
FT misc_RNA 21..205
FT /standard_name="Internal transcribed spacer 1 (ITS1)"
FT rRNA 206..>237
FT /product="5.8S ribosomal RNA"
SQ Sequence 237 BP; 41 A; 77 C; 67 G; 52 T; 0 other;
aacctgcgga aggatcatta ccgagtgcgg gtcctttggg cccaacctcc catccgtgtc 60
tattgtaccc tgttgcttcg gcgggcccgc cgcttgtcgg ccgccggggg ggcgcctctg 120
ccccccgggc ccgtgcccgc cggagacccc aacacgaaca ctgtctgaaa gcgtgcagtc 180
tgagttgatt gaatgcaatc agttaaaact ttcaacaatg gatctcttgg ttccggc 237
//
EMBL/Swiss Prot (http://www.ebi.ac.uk/help/formats_frame.html)
•A sequence in Fasta format begins with a single-line description,
•followed by lines of sequence data.
•The description line is distinguished from the sequence data by a greater-
than (">") symbol in the first column.
•It is recommended that all lines of text be shorter than 80 characters in
length.
>U03518 Aspergillus awamori internal transcribed spacer 1 (ITS1)
AACCTGCGGAAGGATCATTACCGAGTGCGGGTCCTTTGGGCCCAACCTCCCATCCGTGTCTATTGTACCC
TGTTGCTTCGGCGGGCCCGCCGCTTGTCGGCCGCCGGGGGGGCGCCTCTGCCCCCCGGGCCCGTGCCCGC
CGGAGACCCCAACACGAACACTGTCTGAAAGCGTGCAGTCTGAGTTGATTGAATGCAATCAGTTAAAACT
TTCAACAATGGATCTCTTGGTTCCGGC
FASTA
•Exactly one sequence
•Begins with annotation lines
•Start of the sequence is marked by a line ending with "..“
•This line also contains the sequence identifier, the sequence length
and a checksum
ID AA03518 standard; DNA; FUN; 237 BP.
XX
AC U03518;
XX
DE Aspergillus awamori internal transcribed spacer 1 (ITS1) and 18S
DE rRNA and 5.8S rRNA genes, partial sequence.
XX
SQ Sequence 237 BP; 41 A; 77 C; 67 G; 52 T; 0 other; AA03518 Length: 237 Check: 4514
..
1 aacctgcgga aggatcatta ccgagtgcgg gtcctttggg cccaacctcc catccgtgtc
61 tattgtaccc tgttgcttcg gcgggcccgc cgcttgtcgg ccgccggggg ggcgcctctg
121 ccccccgggc ccgtgcccgc cggagacccc aacacgaaca ctgtctgaaa gcgtgcagtc
181 tgagttgatt gaatgcaatc agttaaaact ttcaacaatg gatctcttgg ttccggc
GCG
GenBank/GenPept The nucleotide (GenBank) and protein (Gen Pept) database entries
are available from Entrez in this format
•Can contain several sequences•One sequence starts with: “LOCUS”•The sequence starts with: "ORIGIN“•The sequence ends with: "//“
LOCUS AAU03518 237 bp DNA PLN 04-FEB-1995
DEFINITION Aspergillus awamori internal transcribed spacer 1 (ITS1) and
18S
rRNA and 5.8S rRNA genes, partial sequence.
ACCESSION U03518
BASE COUNT 41 a 77 c 67 g 52 t
ORIGIN
1 aacctgcgga aggatcatta ccgagtgcgg gtcctttggg cccaacctcc catccgtgtc
61 tattgtaccc tgttgcttcg gcgggcccgc cgcttgtcgg ccgccggggg ggcgcctctg
121 ccccccgggc ccgtgcccgc cggagacccc aacacgaaca ctgtctgaaa gcgtgcagtc
181 tgagttgatt gaatgcaatc agttaaaact ttcaacaatg gatctcttgg ttccggc
//
Phylip format
2 2000
G019uabh ATACATCATA ACACTACTTC CTACCCATAA GCTCCTTTTA ACTTGTTAAA
G028uaah CATAAGCTCC TTTTAACTTG TTAAAGTCTT GCTTGAATTA AAGACTTGTT
GTCTTGCTTG AATTAAAGAC TTGTTTAAAC ACAAAAATTT AGAGTTTTAC
TAAACACAAA ATTTAGACTT TTACTCAACA AAAGTGATTG ATTGATTGAT
TCAACAAAAG TGATTGATTG ATTGATTGAT TGATTGATGG TTTACAGTAG
TGATTGATTG ATGGTTTACA GTAGGACTTC ATTCTAGTCA TTATAGCTGC
• The first line of the input file contains the number of sequences and their length (all should have the same length) separated by blanks.
• The next line contains a sequence name, next lines are the sequence itself in blocks of 10 characters. Then follow rest of sequences.
Other formats
MEGA• #mega
• Title: infile.fasta
•
• #G019uabh
• ATACATCATAACACTACTTCCTACCCATAAGCTCCTTTTAACTTGTTAAAGTCTTGCTTG
• AATTAAAGACTTGTTTAAACACAAAAATTTAGAGTTTTACTCAACAAAAGTGATTGATTG
• ATTGATTGATTGATTGATGGTTTACAGTAGGACTTCATTCTAGTCATTATAGCTGCTGGC
• AGTATAACTGGCCAGCCTTTAATACATTGCTGCTTAGAGTCAAAGCATGTACTTAGAGTT
• GGTATGATTTATCTTTTTGGTCTTCTATAGCCTCCTTCCCCATCCCCATCAGTCTTAATC
• AGTCTTGTTACGTTATGACTAATCTTTGGGGATTGTGCAGAATGTTATTTTAGATAAGCA
• AAACGAGCAAAATGGGGAGTTACTTATATTTCTTTAAAGC
• #G028uaah
• CATAAGCTCCTTTTAACTTGTTAAAGTCTTGCTTGAATTAAAGACTTGTTTAAACACAAA
• ATTTAGACTTTTACTCAACAAAAGTGATTGATTGATTGATTGATTGATTGATGGTTTACA
• GTAGGACTTCATTCTAGTCATTATAGCTGCTGGCAGTATAACTGGCCAGCCTTTAATACA
• TTGCTGCTTAGAGTCAAAGCATGTACTTAGAGTTGGTATGATTTATCTTTTTGGTCTTCT
• ATAGCCTCCTTCCCCATCCCATCAGTCT
Don Gilbert
[email protected], May 2001
Indiana University, Bloomington, Indiana
ReadSeq
Seqret
A program in EMBOSS suite
http://www.ebi.ac.uk/cgi-bin/readseq.cgi
http://bioportal.bic.nus.edu.sg/readseq/readseq.html
http://www-bimas.cit.nih.gov/molbio/readseq/
WWW
The Readseq package can read most common formats: examples of all
these formats are included in the readseq directory. The formats include:
• IG/Stanford, used by Intelligenetics and others
• GenBank/GB, genbank flatfile format
• NBRF format (SAM modifications cause this to break when sequences do not have a terminating asterix)
• EMBL, EMBL flatfile format
• GCG, single sequence format of GCG software
• DNAStrider, for common Mac program
• Fitch format, limited use
• Pearson/Fasta, a common format used by Fasta programs and others
• Zuker format, limited use. Input only.
• Olsen, format printed by Olsen VMS sequence editor. Input only.
• Phylip3.2, sequential format for Phylip programs
• Plain/Raw, sequence data only (no name, document, numbering)
• MSF multi sequence format used by GCG software
• PAUP's multiple sequence (NEXUS) format
• PIR/CODATA format used by PIR
Need for databases in Biology?
• Need for storing and communicating large datasets
has grown.
• Need to disseminate biological information.
• Provide Organized data for analysis friendly
retrieval.
• Need to make biological data available in computer-
readable form.
Different classifications of databases
• Type of data
– nucleotide sequences
– protein sequences
– proteins sequence patterns or motifs
– macromolecular 3D structure
– gene expression data
– metabolic pathways
– proteomics data
Different classifications of databases….
• Primary or derived databases
– Primary databases: experimental results
directly into database
– Secondary databases: results of analysis of
primary databases
– Aggregate of many databases
• Links to other data items
• Combination of data
• Consolidation of data
Different classifications of databases….
• Technical design
– Flat-files
– Relational database (SQL)
– Exchange/publication technologies (HTML,
CORBA, XML,...)
• Each one of the above are inter
convertible
Different classifications of databases….
• Availability
– Publicly available, no restrictions
– Available, but with copyright
– Accessible, but not downloadable
– Academic, but not freely available
– Proprietary, commercial; possibly free for
academics
Different classifications of databases….
• Content– Protein/DNA/RNA/miRNA etc.
– Family: kinases
– Common physical properties: membrane bound,
mitochondrial proteins
– Common chemical properties: Proteases, reductases
etc.
– Sequences of a particular genome/species: e.g.
Influenza sequences, plasmodium sequences etc.
– Motifs/domains
Where to look for databases?
• Search Engines
• Journals related to Bioinformatics
• Websites like:– http://www.biophys.uni-duesseldorf.de/BioNet/Pedro/rt_all.html
– www.expasy.ch
– Several others websites
NAR DB issue 2010
• 58 new dbs since last year!
• Total >1230!
• (http://www.oxfordjournals.org/nar/database/a/
• Complete list
– Searchable
– http://nar.oxfordjournals.org/cgi/content/full/gkm1037/DC1/1 (html format), also as downloadable word file)
Database searching tips
• Look for links to Help or Examples
• Always check update dates
• Level of curation
• Try Boolean searches
• Be careful with UK/US spelling differences
– leukaemia vs leukemia
– haemoglobin vs hemoglobin
– colour vs color