pUC18-pUC19-map.pdf
-
Upload
adlina-panca -
Category
Documents
-
view
212 -
download
0
Transcript of pUC18-pUC19-map.pdf
7/27/2019 pUC18-pUC19-map.pdf
http://slidepdf.com/reader/full/puc18-puc19-mappdf 1/2
Reerence
1. Yanisch-Perron, C., et al., Improved M13 phage cloning
vectors and host strains: nucleotide sequences o the
M13mp18 and pUC19 vectors, Gene, 33, 103-119, 1985.
pUC18, pUC19
Enzymes which cut pUC18 DNA once:
AatII 2617Acc65I* 438
AIII 806BamHI* 429BcgI 2215BsaXI 659BstAPI 179BveI* 413CaiI 1217Cr9I* 434Cr10I 1779Eam1105I 1694Ecl136II* 444Eco24I* 444Eco31I 1766Eco88I* 434EcoO109I 2674EcoRI* 450
EheI 235GsuI 1784HincII* 417
HindIII* 399KpnI* 438LguI 683NdeI 183NmeAIII 1822PaeI* 405PdmI 2294PoI 46PscI 806PspFI 1110PstI* 411SacI* 444SalI* 417ScaI 2177SdaI* 410SmaI* 434SspI 2501SspDI 235
XapI* 450XbaI* 423XmiI* 417
* MCS
pUC18 and pUC19 vectors are small, high copy
number, E.coli plasmids, 2686 bp in length. They are
identical except that they contain multiple cloning sites
(MCS) arranged in opposite orientations. pUC18/19
plasmids contain: (1) the pMB1 replicon rep responsibleor the replication o the plasmid (source – plasmid
pBR322). The high copy number o pUC plasmids is
a result o the lack o the rop gene and a single point
mutation in the replicon rep o pMB1; (2) the bla
gene, coding or β-lactamase, that coners resistance
to ampicillin (source – plasmid pBR322). It diers
rom that o pBR322 by two point mutations; (3) the
region o E.coli lac operon containing a CAP protein
binding site, promoter Plac, lac repressor binding site
and the 5’-terminal part o the lacZ gene encoding
the N-terminal ragment o β-galactosidase (source –
M13mp18/19). This ragment, whose synthesis
can be induced by IPTG, is capable o intra-allelic
( α ) complementation with a deective orm o
β-galactosidase encoded by the host (mutation ∆(lacZ)
M15 ). In the presence o IPTG, bacteria synthesize
both ragments o the enzyme and orm blue colonies
on media with X-Gal. Insertion o DNA into the MCSlocated within the lacZ gene (codons 6-7 o lacZ are
replaced by MCS) inactivates the N-terminal ragment
o β-galactosidase and abolishes α-complementation.
Bacteria carrying recombinant plasmids thereore give
rise to white colonies.
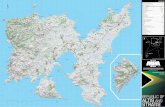
The map shows enzymes that cut pUC18/19 DNA
once. Thermo Scientifc enzymes are shown in orange.
The coordinates reer to the position o the frst
nucleotide in each recognition sequence.
The exact positions o the genetic elements are shown
on the map (termination codons included). The bla
gene nucleotides 2486-2418 (complementary strand)
code or a signal peptide. The LacZ polypeptide
corresponding to wt β-galactosidase and essential or
blue/white screening ends at nt position 236 (compl.
strand); another 30 codons in the same reading rame
are derived rom pBR322. The indicated rep region
is sufcient to promote replication. DNA replicationinitiates at position 866 (+/- 1) and proceeds in the
direction indicated. Plasmids carrying the pMB1 and
ColE1 replicons are incompatible, but they are ully
compatible with those carrying the p15A replicon
(pACYC177, pACYC184). pMB1-derived plasmids can
be amplifed using chloramphenicol.
GenBank/EMBL Accession Numbers
For pUC18 – L09136;
or pUC19 – L09137.
Additional Inormation• CAPproteinbindingsite–591-554(compl.strand);
• mRNA(LacZ)startsatntposition507(compl.strand);
• lac repressor binding site – 507-487 (compl. strand).
Multiple Cloning Site o pUC18
M13/pUC sequencing primer, 17-mer (-20), (#S0100) PstI HincII Cr9I Ecl136II
SdaI SalI Eco88I Acc65I Eco24I EcoRI
399 HindIII PaeI BveI XmiI XbaI BamHI SmaI KpnI SacI XapI 455
5’ - G TAA AAC GAC GGC CAG TGC C AA GCT TGC ATG CCT GCA GGT CGA CTC TAG AGG ATC CCC GGG TAC CGA GCT CGA ATT CGT AAT CAT GGT CAT AGC TGT TTC CTG - 3’
3’ - C ATT TTG CTG CCG GTC A CG GTT CGA ACG TAC GGA CGT CCA GCT GAG ATC TCC TAG GGG CCC ATG GCT CGA GCT TAA GCA TTA GTA CCA GTA TCG ACA AAG GAC - 5’
Val Val Ala Leu Ala Leu Ser Ala His Arg Cys Thr Ser Glu Leu Pro Asp Gly Pro Val Ser Ser Ser Asn Thr Ile Met Thr Met
M13/pUC reverce sequencing primer, 17-mer (-26), (#S0101)
Multiple Cloning Site o pUC19
M13/pUC sequencing primer, 17-mer (-20), (#S0100) Ecl136II Cr9I HincII PstI
EcoRI Eco24I Acc65I Eco88I SalI BveI
396 XapI SacI KpnI SmaI BamHI XbaI XmiI SdaI PaeI HindIII 452
5’ - G TAA AAC GAC GGC CAG TGA ATT CGA GCT CGG TAC CCG GGG ATC CTC TAG AGT CGA CCT GCA GGC ATG CAA GCT TGG CGT AAT CAT GGT CAT AGC TGT TTC CTG - 3’
3’ - C ATT TTG CTG CCG GTC A CT TAA GCT CGA GCC ATG GGC CCC TAG GAG ATC TCA GCT GGA CGT CCG TAC GTT CGA A CC CGA TTA GTA CCA GTA TCG ACA AAG GAC - 5’
Val Val Ala Leu Ala Leu Ser Ala His Arg Cys Thr Ser Glu Leu Pro Asp Gly Pro Val Ser Ser Ser Asn Thr Ile Met Thr Met
M13/pUC reverce sequencing primer, 17-mer (-26), (#S0101)
7/27/2019 pUC18-pUC19-map.pdf
http://slidepdf.com/reader/full/puc18-puc19-mappdf 2/2
There are no restriction sites in pUC18 DNA or the ollowing enzymes:
AanI, AarI, AbsI, AdeI, AjiI, AjuI, AlI, AloI, ApaI, BaeI, BbvCI, BclI, BcuI,
BglII, BoxI, BpiI, BplI, Bpu10I, Bpu1102I, BseJI, BseRI, BsgI, BshTI, Bsp68I,Bsp119I, Bsp120I, Bsp1407I, BspOI, BspTI, Bst1107I, BstXI, Bsu15I, BtgI,
BtgZI, Cr42I, CpoI, CspCI, Eco32I, Eco47III, Eco52I, Eco72I, Eco81I, Eco91I,
Eco105I, Eco130I, Eco147I, FalI, FaqI, FseI, FspAI, Kpn2I, KspAI, MlsI, MluI,
Mph1103I, MreI, MssI, MunI, Mva1269I, NcoI, NheI, NotI, OliI, PacI, PasI,
PauI, PdiI, P23II, Ppu21I, Psp5II, PspXI, PsrI, PsyI, SanDI, SexAI, SaAI, SfI,
SgI, SgrAI, SgrDI, SgsI, SmiI, SrI, Van91I, XagI, XcmI, XhoI, XmaJI.
Indicates restriction sites o Thermo Scientifc enzymes that are sensitive (cleavage completely
blocked or partially inhibited) to overlapping Dam/Dcm methylation. Complete cleavage is achieved with DNA
substrates isolated rom dam – or dcm – hosts.
EnzymeNumber o recognition sites
1 2 3 4 5 6 7 8 9 10 11 12
DrdI 91 908
AatII 2617
Acc65I 438
AIII 806
Alw21I 177 444 1120 2281 2366
Alw26I 51 1766 2531 2684
Alw44I 177 1120 2366BamHI 429
BauI 979 2363 2670
BccI 1735 1859 2146
BceAI 387 1292
BcgI 2215
BcnI 47 82 434 435 1185 1881 2232
BmrI 364 1744
BmI 411 1071 1262 1940
BuI 1015 2542
BglI 245 1813
Bme1390I 47 82 354 434 435 545 833 954 967 1185 1881 2232
BpuEI 912 1174 1451 2319
BsaWI 1012 1159 1990
BsaXI 659
BseDI 354 433 434 545 966
BseGI 77 321 1679 1860 2147
BseLI 47 648 822 840 1006 1285
BseMI 1753 1935
BseMII 171 1081 1490 1656 2196
BseNI 365 391 606 1209 1222 1339 1745 1863 1906 2170 2345
BseSI 177 1120 2366BseXI 41 254 327 630 711 729 1148 1213 1216 1422 1750 2116
Bsh1236I 2 4 107 652 654 852 1433 1763 2256 2588
Bsh1285I 276 719 1143 2066 2215
BshNI 235 438 550 1647
BspLI 235 429 438 550 836 875 1647 1741 1782 1993 2583
BspPI 429 430 1373 1447 1459 1544 1557 2021 2324 2342
BstAPI 179
BsuRI 287 389 646 820 831 849 1283 1741 1821 2088 2675
BtsI 593 2092 2120
BveI 413
CaiI 1217
EaeI 388 645 2087
Cr9I 434
Cr10I 1779
Cr13I 286 1741 1820 1837 2059 2675
CseI 108 908 1486 2236
Csp6I 168 439 2178
DraI 1563 1582 2274
Eam1104I 290 684 2488
Eam1105I 1694
EciI 878 1024 1852
Ecl136II 444
Eco24I 444
Eco31I 1766
Eco47I 1837 2059
Eco57I 1333 2381
Eco88I 434
EcoO109I 2674
EcoP15I 1214 1330 1423
EcoRI 450
EcoRII 354 545 833 954 967
EheI 235
Esp3I 51 2683
FokI 77 321 1679 1860 2147
FspBI 424 1301 1554 1889
GsuI 1784
HaeII 235 680 1050
EnzymeNumber o recognition sites
1 2 3 4 5 6 7 8 9 10 11 12
Hin1I 235 2235 2617
Hin1II 38 406 461 807 1527 2018 2028 2106 2142 2535 2640
HincII 417
HindIII 399
HinI 420 641 706 781 1177 1694
HphI 12 21 1550 1777 2173 2399 2414
Hpy8I 177 417 1120 1608 2366Hpy99I 372 385 907 1701 1964
Hpy188I 31 156 918 996 1349 1483 1618 2064 2075 2195
HpyAV 270 1036 1304 1346 1817 2054 2429
HpyF3I 171 1081 1490 1656 2196 2622
KpnI 438
LguI 683
Lsp1109I 41 254 327 630 711 729 1148 1213 1216 1422 1750 2116
LweI 78 153 208 229 894 1946 2156 2386
MaeIII 57 348 368 1163 1226 1342 1625 1956 2014 2167 2355
MbiI 496 737 2538
MboII 291 685 1456 1547 2302 2380 2489
MmeI 996 1180
MspA1I 112 306 628 1146 1391 2332
MvaI 354 545 833 954 967
NdeI 183
NmeAIII 1822
NmuCI 57 368 1956 2167
NsbI 256 1919
PaeI 405
PagI 1526 2534 2639
PdmI 2294PeI 641 781
PoI 46
PscI 806
Psp1406I 1924 2297
PspFI 1110
PstI 411
PsuI 429 1447 1458 1544 1556 2324 2341
PvuI 276 2066
PvuII 306 628
RsaI 168 439 2178
RseI 1947 2106 2465
SacI 444
SalI 417
ScaI 2177
SchI 420 706 1177 1694
SdaI 410
SduI 177 444 1120 2281 2366
SmaI 434
SmoI 912 1174 1451 2319
FauI 114 124 284 598 655
SspI 2501
SspDI 235
TaaI 19 54 262 768 839 1309 1622 2137
TaiI 374 1509 1925 2298 2618
TaqI 418 448 906 2350
TasI 451 487 504 579 1567 1873 2128
TatI 167 2177
TauI 150 732 850 1005 2089 2211 2440
TscAI 392 593 702 1208 1221 1492 1641 1746 2093 2120
TseI 41 254 327 630 711 729 1148 1213 1216 1422 1750 2116
TspDTI 639 1575 1677 1980
TspGWI 2149 2466
VspI 576 635 1870
XapI 450
XbaI 423
XceI 37 405 806
XmiI 417
pUC18 DNA Restriction Sites