Protein synthesis
-
Upload
nidhiinjbp -
Category
Technology
-
view
1.251 -
download
1
description
Transcript of Protein synthesis

Page 1
Protein synthesis
Made By-
Dr. Nidhi Sharma

Page 2
•Transcription•Genetic code•tRNA•Ribosomes•Translation•Polyribosomes

Page 3
Transcription

Page 4
• Transcription, or RNA synthesis, is theprocess of creating an equivalent RNA copyof a sequence of DNA.
• Both RNA and DNA are nucleic acids, whichuse base pairs of nucleotides as acomplementary language that can beconverted back and forth from DNA to RNAin the presence of the correct enzymes.

Page 5
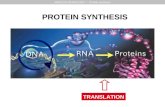
Central dogma
DNA RNA
Proteins
Transcription Translation

Page 6
• During transcription, a DNA sequence isread by RNA polymerase, which produces acomplementary, antiparallel RNA strand.
• As opposed to DNA replication,transcription results in an RNA complimentthat includes uracil (U) in all instanceswhere thymine (T) would have occurred ina DNA compliment.

Page 7
• Transcription is the first step leading to geneexpression.
• The stretch of DNA transcribed into an RNAmolecule is called a transcription unit andencodes at least one gene.
• If the gene transcribed encodes for a protein,the result of transcription is messenger RNA(mRNA), which will then be used to createthat protein via the process of translation.

Page 8
• Alternatively, the transcribed gene mayencode for either ribosomal RNA (rRNA) ortransfer RNA (tRNA), other components ofthe protein-assembly process, or otherribozymes.

Page 9
A DNA transcription unit encoding for a protein containsnot only the sequence that will eventually be directlytranslated into the protein (the coding sequence) butalso regulatory sequences that direct and regulate thesynthesis of that protein.

Page 10
• As in DNA replication, DNA is read from 3' → 5'during transcription. Meanwhile, the complementaryRNA is created from the 5' → 3' direction.
• Only one of the two DNA strands, called the templatestrand, is used for transcription.
• The other DNA strand is called the coding strand,because its sequence is the same as the newlycreated RNA transcript (except for the substitution ofuracil for thymine).

Page 11
• Transcription is divided into 3
stages:
1. initiation
2. elongation
3. termination.

Page 12
Initiation
• In bacteria, transcription begins with the binding of RNApolymerase to the promoter in DNA.
• RNA polymerase is a core enzyme consisting of fivesubunits: 2 α subunits, 1 β subunit, 1 β' subunit, and 1 ωsubunit.
• At the start of initiation, the core enzyme is associatedwith a sigma factor that aids in finding the appropriate -35and -10 base pairs downstream of promoter sequences.

Page 13
Elongation
• One strand of DNA, the template strand (or noncodingstrand), is used as a template for RNA synthesis.
• As transcription proceeds, RNA polymerase traversesthe template strand and uses base pairingcomplementarity with the DNA template to create anRNA copy.
• Although RNA polymerase traverses the templatestrand from 3' → 5', the coding (non-template) strandand newly-formed RNA can also be used as referencepoints, so transcription can be described as occurring5' → 3'.

Page 14
• This produces an RNA molecule from 5' → 3', an exactcopy of the coding strand (except that thymines arereplaced with uracils, and the nucleotides arecomposed of a ribose (5-carbon) sugar where DNA hasdeoxyribose (one less oxygen atom) in its sugar-phosphate backbone).
• Unlike DNA replication, mRNA transcription caninvolve multiple RNA polymerases on a single DNAtemplate and multiple rounds of transcription(amplification of particular mRNA), so many mRNAmolecules can be rapidly produced from a single copyof a gene.

Page 15
• Elongation also involves a proofreading mechanism that can replace incorrectly incorporated bases.

Page 16
Termination
• Bacteria use two different strategies for transcriptiontermination: Rho-independent and Rho-dependent

Page 17
Fig. Formation of
hairpin
In Rho-independenttranscriptiontermination, RNAtranscription stopswhen the newlysynthesized RNAmolecule forms a G-Crich hairpin loopfollowed by a run ofU's, which makes itdetach from the DNAtemplate.

Page 18
Fig. Rho dependent
termination
In the "Rho-dependent"type of termination, aprotein factor called"Rho" destabilizes theinteraction betweenthe template and themRNA, thus releasingthe newly synthesizedmRNA from theelongation complex.

Page 19
Genetic code

Page 20
Properties of genetic code
• The code is universal. All prokaryotic andeukaryotic organisms use the same codon tospecify each amino acid.
• The code is triplet. Three nucleotides make onecodon. 61 of them code for amino acids and 3 viz.,UAA, UAG and UGA are nonsense codons or chaintermination codons.
• The code is degenerate. For a particular aminoacid more than one word can be used

Page 21
• The code is non overlapping. A base in mRNA is not used for two different codons
• The code is commaless. There is no special signal or commas between codons.
• The code is non ambiguous. A particular codonwill always code for the same amino acid, wherever it is found.

Page 22

Page 23
Fig. Base sequence
of yeast alanyl tRNA
Fig. Common features of
tRNA molecule
tRNA

Page 24
Fig. 3D structure of tRNA

Page 25
Ribosomes
• Bacterial ribosomes consists of two subunits ofunequal size, the larger having a sedimentationcoefficient of 50S and the smaller of 30S.
• The two ribosomal subunits have irregularshapes which fit together in such a way that acleft is formed through which mRNA passes asthe ribosome moves along it during thetranslation process and from which the newlyformed polypeptide chain emerges.

Page 26
Fig. Ribosomes

Page 27
Translation

Page 28
• Translation is the first stage of proteinbiosynthesis (part of the overall process of geneexpression).
• Translation is the production of proteins bydecoding mRNA produced in transcription.
• It occurs in the cytoplasm where the ribosomesare located.
• Ribosomes are made of a small and largesubunit which surrounds the mRNA.

Page 29
• In translation, messenger RNA (mRNA) is decodedto produce a specific polypeptide according to therules specified by the genetic code.
• This uses an mRNA sequence as a template toguide the synthesis of a chain of amino acid thatform a protein.
• Many types of transcribed RNA, such as transferRNA, ribosomal RNA, and small nuclear RNA arenot necessarily translated into an amino acidsequence.

Page 30
• Translation proceeds in four phases:
– activation,
– initiation,
– elongation and
– termination
(all describing the growth of the amino acidchain, or polypeptide that is the productof translation).
Amino acids are brought to ribosomes andassembled into proteins.

Page 31
Activation
• In activation, the correct amino acid is covalentlybonded to the correct transfer RNA (tRNA).
• While this is not technically a step in translation, it isrequired for translation to proceed.
• The amino acid is joined by its carboxyl group to the 3'OH of the tRNA by an ester bond.
• When the tRNA has an amino acid linked to it, it istermed "charged".

Page 32
Fig. Charged tRNA (N-formyl
methionyl tRNA fmet)

Page 33
Initiation
In Prokaryotes initiation requires the large and smallribosome subunits, the mRNA, the initiator tRNA andthree initiation factors (IF1, IF2, IF3) and GTP. Theoverall sequence of the event is as follows
• IF3 bind to the free 30S subunit, this helps to preventthe large subunit binding to it without an mRNAmolecule and forming an inactive ribosome
• IF2 complexed with GTP and IF1 then binds to thesmall subunit. It will assist the charged initiator tRNAto bind.

Page 34
• The 30S subunit attached to an mRNA moleculemaking use of the ribosome binding site (RBS) on themRNA
• The initiator tRNA can then bind to the complex bybase pairing of its anticodon with the AUG codon onmRNA.
• At this point, IF3 can be released, as its role inkeeping the subunits apart and helping the mRNA tobind are complete.
• This complex is called 30S initiation complex

Page 35
• The 50S subunit can now bind, which displace IF1and IF2, and the GTP is hydrolysed in this energyconsuming step.
•This complex is called as 70S initiation complex.

Page 36
Fig. Formation of the 70S initiation complex

Page 37
• The assembled ribosome has two tRNAbinding sites.
• These are called the A and P sites, for aminoacyl and peptidyl sites.
• The A site is where incoming amino acyl tRNAmolecules bind, and the P site is where thegrowing polypeptide chain is usually found.
• These sites are in the cleft of small subunitand contain adjacent codons that are beingtranslated.

Page 38
• One major outcome of initiation is theplacement of the initiator tRNA in the Psite.
• It is the only tRNA that does this, as allother must enter the A site.

Page 39
Fig. Initiation phase of protein synthesis

Page 40
Elongation• With the formation of 70S initiation
complex the elongation cycle canbegin.
• In involves three elongation factors EF-Tu, EF-Ts and EF-G, GTP, charged tRNAand the 70S initiation complex.

Page 41
Elongation is divided into three steps:1. Amino acyl tRNA delivery.• EF-Tu is required to deliver the amino acyl tRNA to
the A site and energy is consumed in this step bythe hydrolysis of GTP.
• The released EF-Tu. GDP complex is regeneratedwith the help of EF-Ts.
• In the EF-Tu EF-Ts exchange cycle EF-Ts displacesthe GDP and subsequently is displaced itself byGTP.
• The resultant EF-Tu.GTP complex is now able tobind another amino acyl tRNA and deliver it to theribosome.
• All amino acyl tRNAs can form this complex withEF-Tu except the initiator tRNA.

Page 42
• Fig. Elongation of polypeptide
chain

Page 43
2. Peptide bond formation.• After aminoacyl-tRNA delivery, the A- and P- sites are
both occupied and the two amino acids that are to bejoined are in close proximity.
• The peptidyl transferase activity of the 50S subunitcan now form a peptide bond between these twoamino acids without the input of any more energy,since energy in the form of ATP was used to chargethe tRNA.

Page 44
• Fig. Peptide bond formation

Page 45
3. Translocation.
• A complex of EF-G (translocase) and GTP binds tothe ribosome and, in an energy consuming step, thedischarged tRNA is ejected from the P-site, thepeptidyl-tRNA is moved from the A-site to the P-site and the mRNA moves by one codon relative toone codon to the ribosome.
• GDP and EF-G are released, the latter beingreusable. A new codon is now present in the vacantA-site.
The cycle is repeated until one of thetermination codons (UAA, UAG and UGA) appear inthe A-site.

Page 46
Fig. Translocation

Page 47
Termination • Termination of the polypeptide happens when the A
site of the ribosome faces a stop codon (UAA, UAG,or UGA).
• When this happens, no tRNA can recognize it, but areleasing factor can recognize nonsense codons andcauses the release of the polypeptide chain.
• The 5' end of the mRNA gives rise to the protein's N-terminus, and the direction of translation cantherefore be stated as N->C.

Page 48
Termination• Termination of polypeptide synthesis is signalled
by one of the three termination codons in themRNA (UAA, UAG and UGA) immediatelyfollowing the last amino acid codon.
• In prokaryotes, once a termination codonoccupies the ribosomal A-site, three terminationor release factors, viz., the protein RF1, RF2 andRF3 contribute to-
The hydrolysis of the terminal peptidyl-tRNAbond.
Release of the free polypeptide and the lastuncharged tRNA from the P-site
The dissociation of the 70S ribosome into its 30Sand 50S subunits

Page 49
• RF1 recognizes the termination codon UAGand UAA and RF2 recognize UGA and UAA.
• Either RF1 or RF2 binds at the terminationcodon and induces peptidyl transferase totransfer the growing peptide chain to awater molecule rather than to anotheramino acid.
• Function of RF3 is not known.

Page 50
Fig. Termination

Page 51
Polyribosomes
• A single strand of mRNA is translatedsimultaneously by many ribosomes, spacedclosely together.
• Such clusters of ribosomes are calledpolysomes or polyribosomes.
• The simultaneous translation of a singlemRNA by many ribosomes allow highlyefficient use of the mRNA

Page 52
Fig. Polyribosomes

Page 53
THANK YOU