Protein sequence search tool: A web-based interactive search engine
Transcript of Protein sequence search tool: A web-based interactive search engine

SCIENTIFIC CORRESPONDENCE
CURRENT SCIENCE, VOL. 78, NO. 5, 10 MARCH 2000550
Protein sequence search tool: A web-based interactive search engine
Protein sequence search tool (PSST) isa web-based interactive search tooldeveloped to extract and analyse theprotein/nucleic acid sequence andotherrelated information from the databaseof protein and nucleic acid structures.A search could be performed using allthe protein structures or in a selectedsub-set of non-homologous proteins. Thebasic sequence information is updatedevery week and hence the results ob-tained using the search tool are up-to-date at any given time. The packagePSST (Version 1.0) is available overthe World Wide Web (www) athttp://pranag.physics.iisc.ernet.in/psst
. The number of available three-dimensional structures of proteins andnucleic acids is increasing rapidly. Thethree-dimensional structures of nearly11,800 proteins and nucleic acids arecurrently available in the public do-main of the Protein Data Bank (PDB)1.This database also carries the se-quence information of all the knownthree-dimensional structures of pro-teins and nucleic acids. Prediction ofthe three-dimensional structure fromthe amino acid sequence is one of thecentral problems to be resolved instructural biology. While trying tounderstand this problem it is importantto recognize structural characteristicsof sequence patterns. In several DNA,
RNA and protein structures similarsequence repeat units are often found,and it is essential to know the correla-tion with respect to three-dimensionalstructures. Moreover, identifying theoccurrence of functionally or structur-ally characteristic sequence motifs invarious protein or nucleic acid struc-tures is useful. Occurrence of a par-ticular sequence pattern in a proteinstructure2 might give insights on thepossibility of the associated functionand one could fine-tune the three-dimensional structure by means ofprotein engineering experiments toincorporate the function. Occurrenceof sequence repeats in protein struc-tures might suggest symmetries in
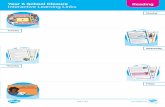
Figure 1. Web page of PSST along with the available search facilities. Clicking on the appropriate search facility activates the search en-gine. The molecule on the left side is a lectin from the seeds of jackfruit. The crystal structure10 of this lectin has four subunits, but only one ofthem is shown. This picture has been created using the graphics software RASMOL11.

SCIENTIFIC CORRESPONDENCE
CURRENT SCIENCE, VOL. 78, NO. 5, 10 MARCH 2000 551
them3. In order to analyse this largevolume of database it is essential tohave automated computational searchtools such as SRS (Sequence RetrievalSystem)4. Towards this effort, severalsoftware packages have been devel-oped for identifying the similarity insequence patterns and their compati-bilities with the available databaseslike SWISS-PROT5 and PIR (ProteinIdentification Resource)6. To the bestof our knowledge, a tool to facilitatesearch for a well-defined molecularweight and also to perform a search forsequence patterns in a set of non-homologous protein sequences is notyet available. PSST addresses theseissues too. The front page of thesearch tool is shown in Figure 1. PSSToffers two different input options forthe user to address her/his query. Theuser has the option of choosing all theprotein structures or only using thenon-homologous protein structuresderived by Hobohm and Sander7. In
this set of non-homologous proteinstructures no two proteins have se-quence identity of over 30%. At pres -ent there are about 1,100 proteinchains in the non-homologous cate-gory. The PSST has the following types ofsearch facility: (a) Sequence retrieval inPIR format, (b) Amino acid composi-tion, (c) sequence length search, (d)molecular weight search, (e) identicalpattern matching and (f) similar patternmatching. The basic input for (a) and(b) is in a four-letter PDB-id code thatusually appears in research papersdealing with the three dimensionalmacromolecular crystal structures. Thesearch tool displays the correspondingresults in an easy and simple format.For example, the amino acid composi-tion search output displays the nameof the amino acid (full name, three-letter code and single-letter code), thenumber of the particular residue and itscontribution towards molecular weight.
It also displays the percentage contentof all the amino acid residues in theinput query protein sequence. At theend of the output it displays the totalmolecular weight of the protein. Thesearch engine requires the lower andupper limits to pick the protein struc-tures, whose lengths satisfy the two-number input (lower and upper bound)criterion for the sequence lengthsearch. Similarly, the molecular weightsearch requires the lower and upperlimit values of the molecular weight inDaltons. A sample output of the user-desired lower and upper limit criterionis shown in Figure 2. The amino acidsequence for the proteins and nucleo-tide sequences for DNA/RNA struc-tures is required as the search input forcarrying out identical/similarity patternmatching. Moreover, PSST has an op-tion of the user-desired number ofmismatches. For example, in the simi-larity search thedefault value for the number of mis -
Figure 2. Typical output of molecular weight search using the lower and upper limits of the user-desired query from the non-homologousprotein sequence database.

SCIENTIFIC CORRESPONDENCE
CURRENT SCIENCE, VOL. 78, NO. 5, 10 MARCH 2000552
matches is 3 and it can be modifiedwhile submitting the web form at theuser end. The identical and similar pat-terns blink with a different colour onthe screen. The results obtained fromthe searches correspond to the mostrecent information available in thePDB1. As mentioned earlier, PSST isavailable on the www and the userscan easily submit their queries. In thetrial runs, the results appear in about1–2 min depending on the networkspeed. A sample output of the result of atypical search for a sequence patternin all the proteins is shown in Figure 3.The pattern searched is taken fromPROSITE8 corresponding to a key
consensus pattern involved in thefunction of phospholipase A2. Thepattern is CCXXHXXC (where C iscysteine, X is any amino acid and H ishistidine) and histidine is the activesite residue. As can be seen from Fig-ure 3 almost all the hits correspond tovarious structures of phospholipaseA 2. This simple search helped in therecognition of all the homologousstructures in a family without per-forming global alignment of completeamino acid sequences. A simple vari-ant of the input sequence patterncould result in the recognition of anon-phospholipase A2. Dependingupon the three-dimensional structureof this
hit one might introduce, by site-directed mutagenesis, the residue nec-essary for the function of phospholi-pase A2 in an otherwise non-phospholipase A2. The sequence and structure data-bases are being updated every weekfrom PDB and fully incorporated withthe PSST search tool. The search en-gine is written in Perl, version 5 (ref. 9)and it can be executed on our NT-4.0Bioinformatics server (a 300 MHzPentium II processor, 128 Mbytes ofmain memory). The front end inputdata part of this tool is written inHTML and JavaScript and allows user-friendly web forms.
1. Bernstein, F. C., Koetzle, T. F., Wil-liams, G. J. B., Meyer, E. F. Jr., Brice,M. D., Rogers, J. R., Kennard, O., Shi-manouchi, T. and Tasumi, M. J., J. Mol.Biol., 1997, 112, 535–542.
2. Kasuya, A. and Thornton, J. M., J. Mol.Biol., 1999, 286, 1673–1691.
3. McLachlan, A. D. and Stewart, M., J.Mol. Biol., 1976, 103, 271–298.
4. Etzold, T., Ulyanov, A. and Argos, P.,Methods Enzymol., 1996, 266, 114–128.
5. Bairoch, A. and Apweiler, R., NucleicAcids Res., 1998, 28, 38–42.
6. Baker, W. C., Garavelli, J. S., Haft,D. H., Hunt, L. T., Marzec, C. R., Or-cutt, B. C., Srinivasarao, G. Y., Yeh, L.S. L., Ledley, R. S., Mewes, H. W.,Pfeiffer, F. and Tsugita, A., NucleicAcids Res., 1998, 28, 27–32.
7. Hobohm, U. and Sander, C., Prot. Sci.,1994, 3, 522–524.
8. Bairoch, A., Bucher, P. and Hofmann,K., Nucleic Acids Res., 1997, 25, 217–221.
9. Wall, L., Christiansen, T. and Schwartz,R. L., Programming Perl, NutshelHandbooks, O'Reilly & Associates Inc.,CA, USA, 1996, 2nd edn.
10. Sankaranarayanan, R., Sekar, K.,Banerjee, R., Sharma, V. Surolia, A. andVijayan., M., Nat. Struct. Biol., 1996, 3,596–602.
11. Sayle, R. A. and Milner-Whilte, E. J.,Trends Biochem. Sci., 1995, 20, 374–382.
ACKNOWLEDGEMENTS. We gratefullyacknowledge the use of the BioinformaticsCentre and the interactive graphics-basedmolecular modelling facility. These facilitiesare supported by the Department of Bio-technology (DBT), Government of India.
Received 29 November 1999; revised ac-cepted 12 January 2000
Figure 3. Sample output frame of the consensus pattern search (only first output frame isshown here). The input search string is CCXXHXXC.

SCIENTIFIC CORRESPONDENCE
CURRENT SCIENCE, VOL. 78, NO. 5, 10 MARCH 2000 553
S. SARAVANAN
A. A JMAL KHAN
K. SEKAR*
Bioinformatics Centre,Raman Building,Indian Institute of Science,Bangalore 560 012, India
*Corresponding author(e–mail: [email protected])