Multiplex digital nucleic acid quantitation using molecular barcodes Paul Rasmussen Sr. Manager of...
-
Upload
priscilla-greaves -
Category
Documents
-
view
222 -
download
1
Transcript of Multiplex digital nucleic acid quantitation using molecular barcodes Paul Rasmussen Sr. Manager of...

Multiplex digital nucleic acid quantitation using molecular barcodes
Paul Rasmussen
Sr. Manager of Emerging Markets and Consumable Programs
NanoString Confidential.1

NanoString Confidential.2
Agenda
Platform Introduction
Chemistry Overview
Performance
Application extensions miRNA and CNVs

NanoString nCounter assay: Single reaction, up to 800 targets
QPCR
Microarrays/NGS
Sens
itivity
Multiplexing # of Transcripts
NanoString
3 NanoString Confidential.
1 10 100 1000 10,000

4
The nCounter platform facilitates powerful research
Over 100 papers have been published using the nCounter platform as of June-2012, at a rate more than doubling yearly
>20% are published in Science, Nature, Cell, or PNAS
Publications span most major disciplines in molecular biology Cancer, Immunology, Stem Cells, Systems Biology, Agriculture
Driven by performance with FFPE and remarkable precisionAccess previously unusable samples Observe biology not previously possible
Publication rate underestimates utilization of the platform by large pharma and industry.
Often less motivated to publish
2008 2009 2010 2011 20120
10
20
30
40
50
60
70
80
90
projected by EOY
pubs per year

The nCounter Analysis System: Two fully automated instruments
Fully Automated sample processing
Up to 800 genes per sample
12 samples processed in one cartridge
Up to 4 cartridges per day
Fully automated imaging and counting
Up to 6 cartridges (72 samples) per day
24 hour unattended processing
Simple data output
5
nCounter Prep Station
nCounter Digital Analyzer

NanoString Confidential.6
Agenda
Platform Introduction
Chemistry Overview
Performance
Application extensions miRNA and CNVs

Each Barcode Attached to an Individual RNA

Two Probe Assay
Target Specific Capture & Reporter Probes are created to bind to the mRNA transcript
Biotin
Target Specific Capture Probe
Target Specific Reporter Probe
8 NanoString Confidential.

Both probes must hybridize
Target Specific Capture & Reporter Probes are created to bind to the mRNA transcript
Target Specific Capture Probe
Target Specific Reporter Probe
9

nCounter CodeSet
Pre-mixed sets of all probes and controls
0
100
200
300
400
500
600
0 20 40 60 80 100 120
Capture Probes
Reporter Probes
System Controls
10 NanoString Confidential.

Day 2AUTOMATED
The nCounter Assay: Three Simple Steps
5 minHANDS-ON
Day 2AUTOMATED
5 minHANDS-ON
Day 15 minHANDS-ON
Purify2
11 NanoString Confidential.
nCounter Prep Station
nCounter Digital Analyzer
Hybridize1
Flexible sample requirements
Only 4 pipetting steps
No amplification
800 hybridizations in single tube
Count3
Sensitive
Precise
Quantitative
Simple

Removeexcessreporters
Bindreporterto surface
Immobilize and align reporter
Image surface
HybridizeCodeSet to RNA
Count codes
nCounter Assay
mRNA Capture & Reporter Probes
12 NanoString Confidential.

Removeexcessreporters
Bindreporterto surface
Immobilize and align reporter
Image surface
HybridizeCodeSet to RNA
Count codes
nCounter Assay
Hybridized mRNA Excess Reporters
13 NanoString Confidential.

Removeexcessreporters
Bindreporterto surface
Immobilize and align reporter
Image surface
HybridizeCodeSet to RNA
Count codes
nCounter Assay
Hybridized Probes Bind to Cartridge
Surface of cartridge is coated with streptavidin
14 NanoString Confidential.

Removeexcessreporters
Bindreporterto surface
Immobilize and align reporter
Image surface
HybridizeCodeSet to RNA
Count codes
nCounter Assay
Immobilize and align reporter for image collecting and barcode counting
15 NanoString Confidential.

Removeexcessreporters
Bindreporterto surface
Immobilize and align reporter
Image surface
HybridizeCodeSet to RNA
Count codes
nCounter Assay
Image Surface
One coded reporter = 1 mRNA
16 NanoString Confidential.

Code Gene Countx 3
y 1
z 2
nCounter Assay
Codes are counted and tabulated
Removeexcessreporters
Bindreporterto surface
Immobilize and align reporter
Image surface
HybridizeCodeSet to RNA
Count codes
17 NanoString Confidential.

NanoString Confidential.18
Simple read out of counts

NanoString Confidential.19
Agenda
Platform Introduction
Chemistry Overview
Performance
Application extensions miRNA and CNVs

The nCounter Assay: Very Reproducible
1
10
100
1000
10000
100000
1 10 100 1000 10000 100000
R2 = 0.9999
Repl
icate
1 C
ount
sReproducibility of NanoString Assay Technical Replicates
Replicate 2 Counts
Data Courtesy of Dr. Roger Bumgarner
20 NanoString Confidential.

Superior Precision in Site-to-Site Reproducibility
Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples
Northcott P.E. et al., Acta Neuropathologica; November 16, 2011
“We present an assay based on NanoString technology that is capable of rapidly, reliably, and reproducibly assigning clinical FFPE medulloblastoma samples to their molecular subgroup, and which is highly suited for future medulloblastoma clinical trials.”
Site 1 Site 2
Site 3
R2 Site1 v Site 2 = 0.97R2 Site1 v Site 3 = 0.98

NanoString® Technologies | Confidential23
Very good cross platform performance: qPCR
Khan et al., 2011

Payton et al, High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. June 2009
Very good cross platform performance: Affymetrix

PCA of signature on Affymetrix and NanoString
Payton et al, High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Invest. June 2009

Very good cross platform performance: RNA-Seq
NanoString Confidential.26
Sun et al. Integrated analysis of Gene Expression, CpG Island Methylation, and Gene Copy Number in Breast Cancer Cells by Deep Sequencing. PLoSone, Feb, 2011

NanoString® Technologies | Confidential27
What samples are you using: flexible options
Total RNA and DNA (100ng/300ng/sample)
Amplified RNA from Small Amount of Sample
LCM and single cell (in progress)
Whole Cell Lysates
PaxGene Lysed Whole Blood
Total RNA and DNA Extracted from FFPE Samples
Crude Extracts from FFPE samples
Plasma, Serum and other Biofluids

Formalin fixation inhibits qPCR much more than the nCounter platform
Heart Frozen/FFPE
0
1
10
100
POLR2a LDHA GRPR C13orf23 NDUFV3
PCRnCounter
Fold
Dec
reas
e

NanoString Confidential.29
Unparalleled Performance on FFPE Samples
mRNA Transcript Quantification in Archival Samples Using Multiplexed, Color-coded ProbesReis, P.P. et al., BMC Biotechnology; May 9, 2011
“… the probe-based NanoString method achieved superior gene expression quantification results when compared to RQ-PCR in archived FFPE samples.We believe that this newly developed technique is optimal for large-scale validation studies using total RNA isolated from archived, FFPE samples.”
nCounter® (r = 0.90) qPCR (r = 0.50)

NanoString Confidential.30
Sample flexibility: Cell Lysate and Matched Total RNA
1 10 100 1000 100000
1
10
100
1000
10000
f(x) = 0.298104770543451 x + 7.194209916585R² = 0.976173007157184
f(x) = 0.630093248384163 x + 12.3939054285541R² = 0.977808883097132
f(x) = 1.30173923776692 x + 22.2432014725715R² = 0.972857263216265
10,000 cells vs 100ngLinear (10,000 cells vs 100ng)5,000 cells vs 100ngLinear (5,000 cells vs 100ng)2,500 cells vs 100ngLinear (2,500 cells vs 100ng)
Counts in 100ng RNA
Coun
ts in
Lys
ate
Measurements with crude whole cell lysates correlate extremely well with purified RNA

NanoString Confidential.31
Flexibility of sample input Total RNA vs Lysate
Khan et al., 2011

NanoString Confidential.32
PaxGene Blood Lysates vs. Purified RNA
-2 0 2 4 6 8 10 12 14 16-2
0
2
4
6
8
10
12
14
16
18
f(x) = 0.985428679843974 x + 0.451779490446374R² = 0.987902256477999f(x) = 0.976041231786931 x + 0.337161048855161R² = 0.983418605725623
Blood Lysate Prep 1Linear (Blood Lysate Prep 1)
log2 counts from total RNA purified from blood
log2
cou
nts
from
blo
od ly
sate
Measurements with unpurified PAXgene blood lysates correlate extremely well with purified RNA

NanoString Confidential.33
Agenda
Platform Introduction
Chemistry Overview
Performance
Application extensions miRNA, and CNVs

34
nCounter miRNA Assays
Human miRNA Panel
Detects:800 human miRNAs3 nonhuman miRNAs (possible spike in controls)
Mouse miRNA Panel
Detects:578 mature mouse miRNAs33 mature murine-associated viral miRNAs
Rat miRNA Panel
Detects:423 mature rat miRNAs
All miRNA Panels
Sample Types Supported: RNA from Fresh/frozen tissue, FFPE, Blood, Cells
Sample Input Recommendation:100 ng purified total RNA
Linear Dynamic Range: 2106 counts
Hands on Time: <2 hours

NanoString Confidential.35
Current challenges to miRNA detection
Short lengthoverall low Tmprohibits concurrent binding, e.g. nCounter dual probe
Highly Related Sequences
Large sequence diversity leads to large Tm spread even though length distribution is fairly small
40 50 60 70 80 900.00%
1.00%
2.00%
3.00%
4.00%
5.00%
6.00%
Human miRNA Tm Distribution
Tm
% H
uman
miR
NA
s

36
miRNA Sample Preparation Basics
miRNAs

37
miRNA Sample Preparation Basics
miRNAs
Hybridize bridge oligoto each miRNA target

38
miRNA Sample Preparation Basics
miRNAs
Bridge oligo specifically annealsto each miRNA target
Unique miRtag for each miRNA species

39
miRNA Sample Preparation Basics
miRNA is covalently linked to miRtag via ligationmiRNAs
Bridge oligo specifically annealsto each miRNA target
Unique miRtag for each miRNA species

miRNA Sample Preparation Basics
Excess bridges and tags are removed
40

Probe Architecture
Target Specific Capture & Reporter Probes bind to the chimaeric miRNA:miRtag molecule
Target Specific Capture Probe
Target Specific Reporter Probe
41
Biotin

NanoString® Technologies | Confidential42
nCounter miRNA Analysis
Multiplexed target profiling of miRNA transcriptomes in a single reaction
Available for human, mouse, rat and drosophila
High level of sensitivity, specificity, precision, and linearity
nCounter®
miRNA AnalysisUnambiguous Discrimination of miRNAs with
Single Nucleotide Differences

Dynamic range of the miRNA assay

Dynamic range of the miRNA assay
hsa-miR-1 expression in different tissues

New miRNA assays for the nCoutner assay
A la carte miRNAsSelect from 20-50 miRNAs from our panels for focused profilingWorkflow identical to standard miRNA assayUsers specify housekeepers (stable miRNAs for normalization)
miRGE assayMixed mRNA and miRNA codesets10-30 miRNAs from our panelsUp to 200 mRNAsWorkflow similar to our standard miRNA assay
NanoString Confidential.45
10 100 1000 1000010
100
1000
10000
R² = 0.988600727780784
GEx vs miRGE assay format probe comparison
Counts for GEx format probes
Cou
nts
for m
iRG
E fo
rmat
pro
bes

NanoString Confidential.46
nCounter Copy Number Assay – Sample Preparation
NanoString Confidential.46
Genomic DNA Fragment ds genomic DNA using 4 base cutter (Alu 1)
Average~ 500 bases
Perform nCounter
Hybridization
Denature DNA @ 95C

Variable copies of X chromosome
Feasibility initially demonstrated with cell lines containing 1, 2, 3, 4 and 5 X chromosomes
Requires fragmentation and denaturation of genomic DNA
The accuracy (measured vs. expected values) obtained with the nCounter system is extremely high
47 NanoString Confidential.

NanoString Confidential.48
Calls agree with HapMap
One copy No copies
No copies
HapMap CNV nCounter CNV

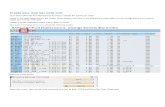
NanoString Confidential.49
Experiment: 100 HapMap samples + reference assayed (600ng digest/300ng hyb)
20 CNV regions with 3 probes analyzed.
Metric % of total Data points
Call rate 94.8% 1902/2006
Accuracy (concordance) 94.2% 1791/1902
Very good correlation with HapMap

NanoString Confidential.50
The importance of Karyotyping….
Normal Female
HeLa