Figure 2 | 3'–5' interactions: circles of mRNA. a | Visualization of circular RNA–protein...
-
Upload
amedeo-bertini -
Category
Documents
-
view
242 -
download
1
Transcript of Figure 2 | 3'–5' interactions: circles of mRNA. a | Visualization of circular RNA–protein...

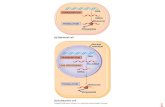
Figure 2 | 3'–5' interactions: circles of mRNA. a | Visualization of circular RNA–protein complexes by atomic-force microscopy. Complexes formed on capped, polyadenylated double-stranded RNA in the presence of eIF4G, poly(A)-binding protein (PABP) and eIF4E91. (Picture provided by A. Sachs and reprinted with permission.) b | Model of messenger-RNA circularization and translational activation by PABP–eIF4G–eIF4E interactions. eIF4G simultaneously binds to eIF4E and PABP7, 9, 14, 53, 55, thereby circularizing the mRNA91 and mediating the synergistic stimulatory effect on translation of the cap and poly(A) tail by enhancing the formation of the 48S complex53, 54, 92. c | Model of mRNA circularization and translational activation by PABP–Paip1 interactions. Paip1 is a PABP-interacting protein that binds eIF4A93, acting as a translational co-activator. d | Model of mRNA circularization and translational repression by CPEB–maskin–eIF4E interactions. RNA-associated CPEB binds maskin, which in turn binds to the eIF4E. This configuration of factors precludes the binding of eIF4G to eIF4E and thus inhibits assembly of the 48S complex13. e | Model of translational repression by heterogeneous nuclear ribonucleoproteins (hnRNPs). The differentiation control element (DICE), located in the 3' UTR of 15-lipoxygenase mRNA, inhibits translation initiation by preventing the joining of the 60S ribosomal subunit to the 43S complex located at the AUG codon. This inhibition is mediated by hnRNP proteins K and E1. The inhibitory event probably targets one of the initiation factors involved in the GTP hydrolysis that releases the initiation factors and the joining of the 60S ribosomal subunit2, 94. ORF, open reading frame.






Ruolo di PABP nella traduzioneRuolo di PABP nella traduzione
In estratti “cell free” di lievito sinergismo tra cap e coda poli(A)
Interazione tra PABP e eIF4G
eIF4E, eIF4G, PABP e mRNA forma strutture circolari (in vitro)
Altre proteine che interagiscono con PABP (Paip1, 2 e eRF3)


Initiation FactorInitiation Factor ActivityActivity
eIF-1 Fidelity of AUG codon recognition
eIF-1A Facilitate Met-tRNAiMet binding to small subunit
eIF-2 Ternary complex formation
eIF-2B (GEF) GTP/GDP exchange during eIF-2 recycling
eIF-3 (10 subunits) Ribosome subunit antiassociation, binding to 40S subunit
eIF-4F (4E, 4A, 4G) mRNA binding to 40S, ATPase-dependent RNA helicase activity
eIF-4A ATPase-dependent RNA helicase
eIF-4E 5' cap recognition
eIF-4G Scaffold for of eIF-4E and -4A in the eIF-4F complex
eIF-4B Stimulates helicase, binds simultaneously with eIF-4F
eIF-4H Similar to eIF4B
eIF-5 Release of eIF-2 and eIF-3, ribosome-dependent GTPase
eIF5B Subunit joining
eIF-6 Ribosome subunit antiassociation

pUp UUUCCUUUU AU G
Inizio di traduzione nell’mRNA di poliovirus
AUG AUG AUG AUG
IRES= Internal ribosome entry site

Saggio dell’mRNA bicistronico
CAT luciferasicap
CAT luciferasicap IRES
CAT luciferasicap IRES
CAT luciferasicap IRES4F
+++ +++
+++ +/-
(+) +++
(+/0) +++




eIF3
40S









CARATTERISTICHE DI UN "SISTEMA VIVENTE"
replicazione
evoluzione

FUNZIONI DELL'RNA NELLA CELLULA MODERNA
traduzione rRNA, mRNA, tRNA
maturazione rRNA snoRNA, RNasi MRP
splicing snRNA, introni gruppi I e II
maturazione tRNA RNasi P
sintesi DNA primers, telomerasi
traslocaz. proteine srpRNA



PERCHE' L'RNA?
l'RNA deve essere venuto prima
del DNA
l'RNA deve essere venuto prima
delle proteine
molti coenzimi hanno un ribonucleotide nella struttura
l'RNA può agire come
catalizzatore