Feed-forward regulation of microbisporicin biosynthesis in ... · 4/8/2011 · 18 mibR , which...
Transcript of Feed-forward regulation of microbisporicin biosynthesis in ... · 4/8/2011 · 18 mibR , which...

1
Feed-forward regulation of microbisporicin 1
biosynthesis in Microbispora corallina 2
3
Lucy Foulston1,2 and Mervyn Bibb1*. 4
5
1 Department of Molecular Microbiology, John Innes Centre, Norwich Research Park, 6
Norwich, Norfolk, NR4 7UH, UK 7
2 Present address: Department of Molecular and Cellular Biology, Harvard University, 8
Cambridge, MA 02138, USA 9
10
11
12
*Corresponding author: 13
Tel: +44-1603-450776; Fax: +44-1603-450778; Email: [email protected] 14
15
Running Title: Regulation of microbisporicin biosynthesis 16
Copyright © 2011, American Society for Microbiology and/or the Listed Authors/Institutions. All Rights Reserved.J. Bacteriol. doi:10.1128/JB.00250-11 JB Accepts, published online ahead of print on 8 April 2011
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

2
Abstract 1
Lantibiotics are ribosomally synthesised, post-translationally-modified peptide 2
antibiotics. Microbisporicin is a potent lantibiotic produced by the actinomycete 3
Microbispora corallina and contains unique chlorinated tryptophan and dihydroxyproline 4
residues. The biosynthetic gene cluster for microbisporicin encodes several putative 5
regulatory proteins including, uniquely, an extracytoplasmic function (ECF) σ factor, 6
σMibX, a likely cognate anti-σ factor, MibW, and a potential helix-turn-helix DNA binding 7
protein, MibR. Here we examine the roles of these proteins in regulating microbisporicin 8
biosynthesis. S1 nuclease protection assays were used to determine transcriptional 9
start sites in the microbisporicin gene cluster and confirmed the presence of likely ECF 10
sigma factor -10 and -35 sequences in five out of six promoters. In contrast, the 11
promoter of mibA, encoding the microbisporicin prepropeptide, has a typical 12
Streptomyces vegetative sigma factor consensus sequence. The ECF sigma factor 13
σMibX was shown to interact with the putative anti-sigma factor MibW in Escherichia coli 14
using bacterial two-hybrid analysis. σMibX auto-regulates its own expression but does not 15
directly regulate expression of mibA. On the basis of quantitative RT-PCR data we 16
propose a model for the biosynthesis of microbisporicin in which MibR functions as an 17
essential master regulator and the ECF sigma factor:anti-sigma factor pair, σMibX:MibW, 18
induces feed-forward biosynthesis of microbisporicin and producer immunity. 19
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

3
Introduction 1
2
Microbisporicin is a potent bactericidal lantibiotic produced by strains of the genus 3
Microbispora (1, 18, 20). In addition to one methyl-lanthionine and three lanthionine 4
bridges, and a C-terminal S-[(Z)-2-aminovinyl]-D-cysteine (1, 18), modifications found in 5
other lantibiotics, microbisporicin contains two unusual modified amino acids: 5-6
chlorotryptophan and 3, 4-dihydroxyproline (1, 18). These modifications are unique to 7
microbisporicin within the lantibiotic class of compounds and are rare in other 8
ribosomally synthesised peptides (although both chlorination and hydroxylation are 9
often found in other natural products (4)). Microbisporicin is active against a wide range 10
of Gram-positive bacterial pathogens, probably through binding to the immediate 11
precursor for cell wall biosynthesis, Lipid II (1, 18). Under the commercial name NAI-12
107, microbisporicin is in preclinical trials and displays superior efficacy in animal 13
models of multi-drug resistant infections compared to the drugs of last resort, linezolid 14
and vancomycin (10). 15
16
A variety of regulatory mechanisms are employed to control lantibiotic production. This 17
may reflect the wide range of input signals that induce lantibiotic biosynthesis in 18
different producing organisms. Many lantibiotics regulate their own production in a cell-19
density dependent manner, frequently mediated by a two-component regulatory system 20
(2), but this is often integrated with other external signals, such as developmental cues, 21
pH and cell stress responses (9, 14, 16, 28). Deletion of mibA, encoding the 22
microbisporicin prepropeptide, from M. corallina NRRL 30420 markedly reduced mib 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

4
gene expression, including that of the putative regulatory genes mibX and mibR 1
suggesting that microbisporicin similarly regulates its own biosynthesis (4). 2
3
Our previous analysis revealed several genes predicted to encode proteins with 4
possible regulatory functions in microbisporicin production including, uniquely for an 5
antibiotic biosynthetic gene cluster, an extracytoplasmic function (ECF) σ factor:anti-σ 6
factor complex (4). σMibX belongs to the extracytoplasmic function (ECF) family of RNA 7
polymerase σ factors, a sub-group that responds to extracellular signals (e.g. cell 8
envelope stress) by influencing transcription initiation through recruitment of RNA 9
polymerase core enzyme at relevant promoter sequences (25). ECF sigma factors differ 10
from other σ70 proteins (the “vegetative” σ factors) in possessing only the conserved σ2 11
and σ4 regions for DNA binding and interaction with RNA polymerase (21), respectively. 12
Furthermore, the consensus motif found at many ECF-dependent promoters differs from 13
that of the σ70 proteins, containing an “AAC” motif in the -35 region and “CGT” 14
nucleotides clustered in the -10 region (8, 17). Many ECF sigma factors auto-regulate 15
their own expression (27). A putative ECF sigma factor consensus motif was identified 16
in five of the six likely promoter regions present in the microbisporicin gene cluster, 17
including that of mibX (Figure 1) (4). 18
19
For the majority of characterised ECF sigma factors a second protein, usually co-20
ordinately expressed, is involved in regulating ECF sigma factor activity. This anti-σ 21
factor is able to sequester the σ factor away from its promoter binding sites, thus 22
preventing transcription initiation until the receipt of a specific signal (27). This signal 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

5
would probably be sensed by the anti-σ factor, or a protein regulating its activity, leading 1
to the release of the σ factor and subsequent expression of its target genes. This 2
release can be mediated via a conformational change (for example, regulation of σR by 3
RsrA in S. coelicolor (25)) or via proteolytic degradation (7). mibW, downstream of and 4
apparently translationally coupled to mibX, encodes a protein predicted to function as 5
an anti-sigma factor protein for σMibX (4). 6
7
We previously suggested a model in which σMibX is required for high-level expression of 8
the microbisporicin gene cluster (including its own gene) and in which its activity 9
depends on low levels of production of microbisporicin (4). We proposed that low levels 10
of microbisporicin (potentially produced by expression from a starvation-induced 11
promoter) or cell envelope stress (induced by the likely interaction of microbisporicin 12
with Lipid II) prevents MibW from interacting with σMibX, thus releasing the ECF σ factor 13
and resulting in high-level expression of the entire microbisporicin gene cluster. 14
Interestingly, the operon beginning with mibA lacks the ECF sigma factor consensus 15
motif, suggesting that regulation by σMibX might be mediated through a gene in one of 16
the other σMibX-regulated operons. We suggested that a likely candidate for this is 17
mibR, which encodes a protein with a predicted helix-turn-helix DNA binding domain 18
between amino acids 173 to 214 (Pfam; (3)). 19
20
In this study, we characterise the functions of σMibX, MibW and MibR and their roles in 21
regulating microbisporicin biosynthesis in M. corallina. 22
23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

6
Materials and Methods 1
Strains and general methods 2
Oligonucleotides and plasmids are described in Table S1. M. corallina NRRL 30420 3
was grown and manipulated, and microbisporicin detected, as described previously (4). 4
High-resolution S1 nuclease mapping 5
RNA was purified from exponential-phase wild type M. corallina as described previously 6
(4). Probes for S1 nuclease protection analysis were generated by PCR using the 7
oligonucleotide pairs shown in Table S1. Non-homologous tails (underlined) were 8
incorporated into some probes to distinguish between full-length protection and 9
probe:probe re-annealing. Asterisks mark the oligonucleotides, internal to the respective 10
protein coding sequences, which were labelled at the 5’ end with 32
P-Υ-ATP. The 11
oligonucleotides were labelled using T4 polynucleotide kinase (Epicentre) following the 12
manufacturer’s instructions. Hybridisations were carried out using 30 µg wild type M. 13
corallina RNA in sodium trichloroacetic acid buffer at 45°C overnight after denaturation 14
at 70°C for 10 min (15). G+A sequencing ladders were generated from the end-labelled 15
probes by chemical sequencing (22). 16
Bacterial two-hybrid analysis 17
pT25 contains a multiple-cloning site (MCS) preceded by a fragment of cya of 18
Bordetella pertussis that encodes half of the catalytic domain (T25) of adenylate cyclase 19
(CyaA) (12). XP461 is pT25 containing the leucine zipper fragment (13). The other half 20
of CyaA (T18) is encoded by pUT18 and is preceded by a MCS. pUT18C is similar to 21
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

7
pUT18 except that the MCS follows the cya gene fragment, allowing the generation of 1
C-terminal rather than N-terminal fusion proteins (13). The leucine zipper fragment from 2
XP461 was removed by KpnI digestion and cloned in the KpnI sites of pUT18 and 3
pUT18C to generate the positive control plasmids pUT18-zip and pUT18C-zip, 4
respectively. 5
mibX was amplified by high-fidelity PCR from pIJ12125 using the primers LF101F and 6
LF101R (no stop codon included), and LF101F and LF101R2 (stop codon included), 7
which introduced BamHI and KpnI sites (5’ and 3’, respectively, with respect to mibX) 8
into the resulting PCR products. The PCR products were gel-purified, digested with 9
BamHI and KpnI, and ligated into pUT18 and pUT18C cut with BamHI and KpnI to 10
generate pIJ12367 and pIJ12368, respectively. mibW was amplified by high-fidelity 11
PCR from pIJ12125 using primers LF102F and LF102R, which introduced BamHI and 12
KpnI sites into the resulting PCR product. The PCR product was gel-purified, digested 13
with BamHI and KpnI, and ligated into XP458 (pT25) cut with BamHI and KpnI to 14
generate pIJ12369. All constructs were confirmed by Sanger sequencing. 15
Combinations of the resulting plasmids were introduced into E. coli BTH101 by co-16
transformation and screened on MacConkey medium (Difco) containing 1% maltose, 17
0.5 mM IPTG, 25 µg/ml chloramphenicol and 100 µg/ml carbenicillin. Plates were 18
incubated at 30°C for several days until the colour of colonies had developed fully. 19
Quantification of β-galactosidase activity was carried out as previously described (26). 20
Two independent clones from each interaction plate were assayed for β-galactosidase 21
activity and the values averaged. 22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

8
Luciferase assays 1
The intergenic region between mibX and mibA, with and without mibX, was amplified by 2
PCR using primer pairs containing EcoRI and BamHI sites, and in both orientations with 3
respect to the restriction sites. The primers used were LF096F and LF096R (pIJ12341), 4
LF096F2 and LF096R (pIJ12342), LF097F and LF097R (pIJ12343), and LF097F and 5
LF097R2 (pIJ12344). The resulting PCR fragments were cloned into EcoRI-BamHI-cut 6
pIJ5972, an integrative, Streptomyces promoter-probe plasmid containing TTA codon-7
free derivatives of the luxAB reporter genes (19). The resulting constructs and pIJ5972 8
(negative control) were transferred by conjugation into S. coelicolor M1146 (5). Plasmid-9
containing strains were grown on Difco Nutrient Agar in single wells of a 25-well plate 10
(10cm x 10cm; Sterilin) for 2 days. Each well was inoculated with approximately 5 × 106 11
spores. Plates were exposed to filter paper impregnated with n-decanal for 5 min and 12
luciferase activities were observed using a NightOwl camera (Berthold) equipped with 13
WinLight software (Berthold) using a 1 min exposure time. 14
Construction of M. corallina deletion mutants 15
Genes in pIJ12125 (4) were replaced with an apramycinR (apr)-oriT cassette amplified 16
from pIJ773 using the primer pairs listed in Table S1, as described in (6). Mutations 17
were confirmed by PCR using flanking primers and by restriction digests to confirm the 18
integrity of the targeted cosmid. Conjugations between E. coli ET12567/pUZ8002 19
carrying the oriT-containing cosmid and M. corallina were carried out as previously 20
described (4). Deletion mutants of M. corallina were constructed as described 21
previously (4). 22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

9
Complementation of ∆mibR::apr 1
To complement the mibR deletion mutant, a PCR product was generated by high-fidelity 2
PCR using primers LF117F and LF112R that contained 32 bp of the 3’ end of mibQ 3
(including the stop codon), the intergenic region between mibQ and mibR (including the 4
promoter PmibR identified by S1 mapping), and mibR. This fragment was blunt-end 5
cloned into the EcoRV site of pIJ10706 (4) to generate pIJ12376, which was confirmed 6
by Sanger sequencing. pIJ12376 and pIJ10706 (the empty vector control) were 7
transferred into the ∆mibR mutant by conjugation from E. coli ET12567/pUZ8002. 8
Quantitative reverse transcriptase PCR 9
M. corallina wild type and mutant strains were cultured in triplicate as described 10
previously (4). Mycelial growth was followed by measuring the optical density at 450 nm 11
of 1 ml of culture taken from each of three replicate cultures at different time points. 12
RNA was extracted from 2.5 ml of mycelium of each sample at 48 h of growth as 13
described previously (4). 2.5 µg RNA were treated with DNase I in a 25-µl volume 14
following the manufacturer’s instructions (Amplification Grade, Invitrogen). 8 µl of each 15
of the resulting RNA samples were converted to cDNA (4). To control for DNA 16
contamination in the qRT-PCR, a duplicate set of cDNA synthesis reactions were 17
performed but with the reverse transcriptase enzyme omitted. Following RNase H 18
treatment, the samples were diluted 1:100 with nuclease-free water and 2.5 µl were 19
used in the quantitative PCR reaction with SYBR Greener qPCR Supermix (Invitrogen) 20
according to the manufacturer's instructions. Each 25 µl reaction contained 200 nM of 21
forward and reverse primers and 5% DMSO. PCR cycling was performed in a Chromo4 22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

10
machine (BioRad, CA, USA) at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 1
95°C for 15 s, 58°C for 60 s and 72°C for 1 s. Parallel reactions were performed in the 2
same 96-well plate using different dilutions of M. corallina genomic DNA to generate a 3
standard curve for each gene analysed. All determinations were performed in triplicate, 4
and the results were analysed using Opticon 2 Monitor software (MJ Research, 5
Waltham, MA, USA). Values were normalized to an endogenous control gene, hrdB, 6
encoding the homolog of the vegetative sigma factor of S. coelicolor (4). All the control 7
samples from cDNA synthesis lacking reverse transcriptase gave values comparable to 8
background, indicating that the RNA samples were not contaminated with genomic 9
DNA. 10
11
Results 12
13
High–resolution S1 nuclease mapping of transcriptional start sites in the 14
microbisporicin gene cluster 15
16
Bioinformatic analysis of the mib gene cluster revealed five likely intergenic regions, one 17
of them divergent. The mib gene cluster was thus predicted to consist of six operons 18
(Figure 1) and ECF-sigma factor consensus motifs were identified upstream of five of 19
them (4). 20
21
32P-labelled probes homologous to the predicted 5’ ends of the transcripts of mibJ, 22
mibQ, mibR, mibX, mibA and mibE were used to map the transcriptional start site of 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

11
each gene by S1 nuclease protection analysis (Figure 2 and Figure S1). S1 nuclease-1
protected fragments were sized by polyacrylamide gel electrophoresis and comparison 2
to DNA size markers. Fragments that could not be accurately sized by this method were 3
further analysed by comparison to a GA-sequencing ladder derived from the respective 4
5’ end-labelled probe (Figure 2 and S1). 5
The transcriptional start sites of mibJ, mibQ, mibR, mibX and mibE lie 8-10 nucleotides 6
downstream of the identified ECF-sigma factor consensus -10 sequences (Figure 3). 7
This suggests that these genes and operons are subject to direct transcriptional control 8
by the ECF-sigma factor σMibX, which is essential for microbisporicin biosynthesis (4). 9
10
Non-homologous sequences at the 5’ end of the probes for mibJ, mibQ and mibR were 11
used to distinguish between transcriptional read-through from upstream promoters and 12
probe:probe re-annealing. Fragments approximately 15 nucleotides shorter than the 13
respective probes for mibQ and mibR indicated the additional presence of 14
transcriptional read-through into these genes (Figure S1). 15
16
The transcriptional start site of mibA was not preceded by an ECF-sigma factor 17
consensus sequence (Figure 4). Instead, -35 and -10 sequences closely matching 18
those of the consensus sequence of the housekeeping sigma factor of Streptomyces 19
coelicolor (29) were identified upstream of the mibA transcriptional start (Figure 4). The 20
predicted -35 element of mibA matches exactly that of several S. coelicolor and E. coli 21
σ70 promoters and the -10 element includes the strictly conserved “T” at position 6 as 22
well as the preferred nucleotides “T” at position 1 and “GA” at positions 3 and 4, 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

12
respectively (29). The spacing of 17 nucleotides between these two elements is also 1
characteristic of this group of promoters (29). This suggests that mibA transcription, 2
unlike that of the other mib genes, is not mediated by σMibX, but probably by the 3
housekeeping sigma factor of M. corallina. However, transcription of mibA might be 4
influenced by other transcriptional regulators acting at the mibA promoter. 5
6
Analysis of the interaction between σMibX and MibW 7
8
MibW was proposed previously to function as an anti-sigma factor that regulated the 9
activity of σMibX (4). MibW would bind to σMibX, possibly through its predicted N-terminal 10
cytoplasmic domain, and thus tether it to the membrane, where MibW would be 11
embedded via its predicted transmembrane helices. To determine if an interaction 12
occurred between MibW and σMibX, a bacterial-two-hybrid (BACTH) experiment was 13
performed. 14
15
mibX was fused to the gene encoding the T18 fragment of adenylate cyclase in two 16
vectors: pUT18, in which σMibX would be at the N-terminus of the fusion protein 17
(pIJ12367), and pUT18C, in which σMibX would be at the C-terminus of the fusion protein 18
(pIJ12368). mibW was introduced into pT25, resulting in fusion of the putative anti-19
sigma factor to the C–terminus of the T25 fragment of adenylate cyclase. pT25 and 20
pUT18 or pUT18C, all containing the leucine zipper fragment (zip) from GCN4 (yeast 21
protein), were used as positive controls. The leucine zipper domain interacts strongly 22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

13
with itself and provides a reliable positive control for bacterial-two hybrid studies (12). 1
Negative controls were the empty vectors. 2
3
Interacting pairs of proteins were screened initially by transforming E. coli BTH101 with 4
the appropriate plasmids and monitoring restoration of adenylate cyclase activity on 5
MacConkey-Maltose indicator agar plates incubated at 30°C for 4 days (12). Interaction 6
was readily observed between T25- MibW and both T18-σMibX and T18C-σMibX (data not 7
shown), indicating that the position of the T18 fragment did not affect the ability of MibW 8
to interact with the ECF sigma factor. Two clones of each interaction pair were assayed 9
for β-galactosidase activity. Interaction between MibW and σMibX was confirmed, with β-10
galactosidase activity higher than that of the leucine-zipper positive control fragments 11
(Figure 5). 12
13
σMibX induces transcription from the mibX promoter in Streptomyces coelicolor 14
15
A series of constructs were derived from the reporter plasmid pIJ5972, which contains a 16
promoterless luxAB cassette encoding the enzyme luciferase and which integrates in 17
the ΦC31 attachment site of the chromosome of S. coelicolor (Figure 6) (19). pIJ12341 18
contained the intergenic region between mibX-mibA (with the mibA promoter driving 19
expression of luxAB) and mibX (with expression driven from its own promoter), whereas 20
pIJ12342 contained only the intergenic region. pIJ12343 contained the intergenic region 21
between mibX-mibA (with the mibX promoter driving expression of luxAB) and mibX, 22
whereas pIJ12344 contained only the intergenic region (Figure 6). These constructs, 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

14
along with pIJ5972, were introduced into S. coelicolor M1146 by conjugation (5). Three 1
independent clones were selected from each conjugation and grown in duplicate for two 2
days on Difco Nutrient Agar (where S. coelicolor does not make aerial hyphae and 3
spores, which can interfere with light emission). The mycelium was exposed for 5 4
minutes to N-decanal that had been spotted onto filter discs and light emission detected 5
using a NightOwl camera. No light production was detected from the vector-only control 6
strain or from strains containing pIJ12341 or pIJ12342, where luxAB were located 7
downstream of the mibA promoter. A very low level of light production was observed 8
from strains containing pIJ12344, indicative of a low level of transcriptional activity from 9
the mibX promoter in the absence of σMibX, possibly mediated by one of the other ECF 10
sigma factors of S. coelicolor (25). In contrast, high levels of light emission were 11
observed from the pIJ12343 clones containing mibX and with the luxAB genes 12
transcribed from the mibX promoter. This suggests that σMibX positively auto-regulates 13
its own expression but does not direct transcription from the mibA promoter. 14
15
mibR is essential for production of microbisporicin in M. corallina 16
17
pIJ12125, containing the entire mib gene cluster but with the apramycin cassette from 18
pIJ773 replacing mibR, was mobilised into M. corallina by conjugation. Two clones were 19
identified that gave rise to single colonies that were apramycin resistant yet sensitive to 20
kanamycin (resistance conferred by the cloning vector), indicating the occurrence of 21
double crossing over. Replacement of mibR in these clones by the apramycin 22
resistance cassette was confirmed by PCR. Since mibR lies in a mono-cistronic operon 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

15
(Figure 1) and is adjacent to the convergently transcribed mibW, its deletion should not 1
have polar effects on the expression of other mib genes. The two clones were grown in 2
VSPA liquid medium (4) for seven days along with the wild type strain. Unlike the wild 3
type control, supernatants from the mutant strains did not generate a zone of inhibition 4
when spotted onto a lawn of Micrococcus luteus (Figure 7; one representative clone 5
shown). Bioactivity and microbisporicin production (confirmed by MALDI-ToF analysis) 6
were restored by complementation in trans, with mibR expressed from its own promoter. 7
Thus mibR is essential for microbisporicin biosynthesis. 8
9
Deleting mibX or mibR affects transcription of the mib gene cluster 10
11
Wild-type M. corallina and the ∆mibX::apr and ∆mibR::apr mutants were grown in 12
triplicate in VSPA liquid medium for RNA isolation and in triplicate for the assessment of 13
mycelial growth by measuring the optical density at 450 nm. The three strains grew 14
initially at similar rates, although the ∆mibX::apr mutant exhibited a slower growth rate 15
at later time points but accumulated to the same optical density in stationary phase as 16
the other strains (Figure S2). Samples of supernatant were taken at intervals to assess 17
microbisporicin biosynthesis by bioassay against M. luteus. The earliest time point at 18
which bioactivity could be detected in the wild type was 40 h (Figure S2). There was no 19
bioactivity with the supernatants from the ∆mibX::apr or ∆mibR::apr mutants even after 20
seven days growth (the latest time point assayed) (data not shown). 21
22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

16
To assess the effect of deleting mibX and mibR on transcription of the mib gene cluster, 1
RNA was isolated from 2.5 ml of mycelium from each of the three replicate cultures of 2
each strain after 48 h growth, i.e. after microbisporicin biosynthesis had begun in the 3
wild type strain. mib gene expression was assessed by quantitative reverse 4
transcriptase (RT) PCR using the M. corallina homolog of hrdB, the vegetative sigma 5
factor gene of S. coelicolor, as an internal control (4). Representative genes were 6
chosen from each predicted operon of the mib cluster and their expression assessed 7
using the primers listed in Table S1. 8
9
Expression of all of the mib genes tested was strongly dependent on both mibX and 10
mibR (Figure 8). Interestingly, mibA and mibD expression levels were 4- and 5.4-fold, 11
respectively, lower in the ∆mibR mutant than in the ∆mibX mutant. Conversely, levels 12
of expression of mibJ, mibQ and mibE were 1.84-, 6.95- and 2.4-fold, respectively, 13
lower in the ∆mibX mutant than in the ∆mibR mutant. When compared to wild type 14
levels, expression of mibR was reduced to a far lesser extent (8.8-fold) in the ∆mibX 15
mutant than mibJ, mibQ or mibE (2561-, 1398- and 3124-fold, respectively). These 16
results are consistent with direct regulation of mibJ, mibQ and mibE by σMibX, and direct 17
regulation of the mibA operon by MibR. 18
19
Discussion 20
21
Regulation of microbisporicin biosynthesis is complex and appears to rely on a strict 22
interplay between three regulatory proteins, MibR, σMibX, MibW, and production of 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

17
microbisporicin itself. S1 nuclease protection analysis confirmed that the previously 1
identified putative ECF sigma factor consensus sequences lie appropriate distances 2
from transcriptional start sites to act as promoter sequences for all of the operons in the 3
mib cluster except mibA. This strongly suggests that σMibX, an ECF sigma factor 4
encoded within the cluster and essential for microbisporicin production, directs 5
transcription of these operons. Furthermore, σMibX directed its own expression in a 6
heterologous host, presumably through recruitment of RNA polymerase to the 7
consensus motif in its own promoter region. 8
9
Interaction between full-length σMibX and MibW was confirmed in E. coli. This is 10
consistent with MibW functioning as a σMibX-specific anti-sigma factor, sequestering it at 11
the membrane (MibW is a predicted transmembrane protein) in the absence of an 12
activating signal. MibW, by regulating σMibX activity, would thus play a crucial role in 13
regulating microbisporicin production. Our attempts to delete mibW in M. corallina have 14
proved unsuccessful, resulting in the isolation of an apparent suppressor mutation in 15
mibX that is likely to abolish sigma factor function (data not shown). Thus, in the 16
absence of MibW, constitutive production of microbisporicin might overwhelm the 17
organism’s immunity system, potentially mediated by MibEF (4), resulting in cell death. 18
Attempts to complement the ∆mibX::apr mutation with mibXW expressed in trans from 19
their native promoter restored microbisporicin production, but not to wild-type levels, 20
potentially reflecting a surfeit of MibW in the complemented strain, and highlighting the 21
importance of a 1:1 stoichiometry for the two proteins for regulation. 22
23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

18
Our results suggest a model for the regulation of microbisporicin biosynthesis in which 1
MibR functions as a master regulator to promote low-level microbisporicin biosynthesis, 2
which subsequently induces a feed-forward mechanism mediated by σMibX that results in 3
high level microbisporicin production (Figure 9). Prior to the onset of detectable 4
microbisporicin production, a basal level of expression of mibXW leaves the system 5
poised for activation, with σMibX sequestered at the membrane. We propose that an 6
unknown signal, possibly nutrient limitation, results in activation of transcription of mibR 7
mediated through a growth rate-dependent promoter. This would result in expression of 8
the mibABCDTUV operon and production of a form of microbisporicin that lacks 9
chlorination of tryptophan at position 4, and hydroxylation of proline at position 14, and 10
which may therefore possess reduced antibiotic activity (18). Export, potentially 11
mediated by MibTU, would permit either direct interaction of the peptide with MibW, or a 12
low level of inhibition of peptidoglycan biosynthesis that may be perceived by the anti-13
sigma factor. Either signal could then result in inactivation of MibW, release of σMibX, 14
and high level expression of the entire mib gene cluster, including genes we predict to 15
confer immunity to microbisporicin, namely those encoding the ABC transporter MibEF 16
(4) and the putative lipoprotein MibQ (LanI lipoproteins confer immunity in other 17
lantibiotic producing organisms (2)). Furthermore induction of expression of mibHS, 18
required for tryptophan chlorination, and of mibO, encoding a cytochrome P450 19
probably involved in proline hydroxylation, would result in the formation of fully 20
processed and active microbisporicin (4). 21
22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

19
Deletion of mibX had a less severe effect on expression of mibA and mibD than deletion 1
of mibR, consistent with direct regulation of mibABCDTUV by MibR and not by σMibX. 2
This is also consistent with the results of the luciferase assays in which mibA 3
expression was not induced by σMibX. Conversely, expression of mibJ, mibQ, mibE was 4
more severely affected by deletion of mibX than deletion of mibR, consistent with their 5
direct regulation by σMibX, and with the presence of the ECF sigma factor consensus 6
sequence upstream of their respective transcriptional start sites. 7
8
Feed-forward regulation of antibiotic biosynthesis, where biosynthetic pathway 9
intermediates as well as the final product are postulated to activate export and 10
potentially immunity mechanisms, was suggested previously for actinorhodin production 11
in Streptomyces coelicolor (30), and other examples also exist (11, 23-24). 12
13
Our results contribute not only to our knowledge of the regulatory mechanisms used to 14
control lantibiotic biosynthesis in actinomycetes but provide a basis for rational attempts 15
to improve the level of microbisporicin production for pharmaceutical development and 16
application by manipulation of the regulatory genes mibR and mibXW. 17
18
Acknowledgements 19
We thank several colleagues at the John Innes Centre: David Hopwood and Mark 20
Buttner for comments on the manuscript; Juan-Pablo Gomez-Escribano, Maureen Bibb, 21
Tung Le, Ngat Tran and Jane Moore for technical advice and assistance; the Dixon and 22
Downie labs for the gift of BACTH plasmids and for technical advice; and Gerhard 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

20
Saalbach and Mike Naldrett for mass spectrometry. This work was supported financially 1
by a Doctoral Training Grant to L.C.F. and by funding to M.J.B., both from the 2
Biotechnology and Biological Sciences Research Council, United Kingdom.3
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

21
1
References 2
3
1. Castiglione, F., A. Lazzarini, L. Carrano, E. Corti, I. Ciciliato, L. Gastaldo, P. 4
Candiani, D. Losi, F. Marinelli, E. Selva, and F. Parenti. 2008. Determining the 5
structure and mode of action of microbisporicin, a potent lantibiotic active against 6
multiresistant pathogens. Chem. Biol. 15:22-31. 7
2. Chatterjee, C., M. Paul, L. Xie, and W. A. van der Donk. 2005. Biosynthesis 8
and mode of action of lantibiotics. Chem. Rev. 105:633-84. 9
3. Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. 10
Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. 11
The Pfam protein families database. Nucleic Acids Res. 36:D281-8. 12
4. Foulston, L. C., and M. J. Bibb. 2010. Microbisporicin gene cluster reveals 13
unusual features of lantibiotic biosynthesis in actinomycetes. Proc. Natl. Acad. 14
Sci. U. S. A. 107:13461-6. 15
5. Gomez-Escribano, J. P., and M. J. Bibb. 2010. Engineering Streptomyces 16
coelicolor for heterologous expression of secondary metabolite gene clusters. 17
Microbial Biotechnol. doi: 10.1111/j.1751-7915.2010.00219.x. 18
6. Gust, B., T. Kieser, and K. F. Chater. 2002. REDIRECT© technology: PCR-19
targeting system in Streptomyces coelicolor. The John Innes Centre, Norwich, 20
United Kingdom. 21
7. Heinrich, J., K. Hein, and T. Wiegert. 2009. Two proteolytic modules are 22
involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. 23
Microbiol. 74:1412-26. 24
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

22
8. Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. 1
Microb. Physiol. 46:47-110. 2
9. Hindre, T., J. P. Le Pennec, D. Haras, and A. Dufour. 2004. Regulation of 3
lantibiotic lacticin 481 production at the transcriptional level by acid pH. FEMS 4
Microbiol. Lett. 231:291-8. 5
10. Jabes, D., C. Brunati, G. Candiani, S. Riva, G. Romano, and S. Donadio. 6
2011. Efficacy of the new lantibiotic NAI-107 in experimental infections induced 7
by MDR Gram positive pathogens. Antimicrob. Agents Chemother. AAC.01288-8
10v1 9
11. Jiang, H., and C. R. Hutchinson. 2006. Feedback regulation of doxorubicin 10
biosynthesis in Streptomyces peucetius. Res. Microbiol. 157:666-74. 11
12. Karimova, G., J. Pidoux, A. Ullmann, and D. Ladant. 1998. A bacterial two-12
hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. 13
Acad. Sci. U. S. A. 95:5752-6. 14
13. Karimova, G., A. Ullmann, and D. Ladant. 2001. Protein-protein interaction 15
between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains 16
revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73-82. 17
14. Kies, S., C. Vuong, M. Hille, A. Peschel, C. Meyer, F. Gotz, and M. Otto. 2003. 18
Control of antimicrobial peptide synthesis by the agr quorum sensing system in 19
Staphylococcus epidermidis: activity of the lantibiotic epidermin is regulated at 20
the level of precursor peptide processing. Peptides 24:329-38. 21
15. Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. 22
Practical Streptomyces Genetics John Innes Foundation, Norwich. 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

23
16. Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. 1
de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal 2
transduction. J. Biol. Chem. 270:27299-304. 3
17. Lane, W. J., and S. A. Darst. 2006. The structural basis for promoter -35 4
element recognition by the group IV sigma factors. PLoS Biol. 4:e269. 5
18. Lazzarini, A., L. Gastaldo, G. Candiani, I. Ciciliato, D. Losi, F. Marinelli, E. 6
Selva, and F. Parenti. February 2005. Antibiotic 107891, its factors A1 and A2, 7
pharmaceutically acceptable salts and compositions, and use thereof. 8
WO/2005/014628. 9
19. Le, T. B., H. P. Fiedler, C. D. den Hengst, S. K. Ahn, A. Maxwell, and M. J. 10
Buttner. 2009. Coupling of the biosynthesis and export of the DNA gyrase 11
inhibitor simocyclinone in Streptomyces antibioticus. Mol. Microbiol. 72:1462-74. 12
20. Lee, M. D. April 2003. Antibiotics from Microbispora. U.S. patent 6551591. 13
21. Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of 14
the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of 15
eubacterial RNA polymerase sigma factors involved in the regulation of 16
extracytoplasmic functions. Proc. Natl. Acad. Sci. U. S. A. 91:7573-7. 17
22. Maxam, A. M., and W. Gilbert. 1980. Sequencing end-labeled DNA with base-18
specific chemical cleavages. Methods Enzymol. 65:499-560. 19
23. Ostash, I., B. Ostash, A. Luzhetskyy, A. Bechthold, S. Walker, and V. 20
Fedorenko. 2008. Coordination of export and glycosylation of landomycins in 21
Streptomyces cyanogenus S136. FEMS Microbiol. Lett. 285:195-202. 22
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

24
24. Otten, S. L., J. Ferguson, and C. R. Hutchinson. 1995. Regulation of 1
daunorubicin production in Streptomyces peucetius by the dnrR2 locus. J. 2
Bacteriol. 177:1216-24. 3
25. Paget, M. S. B., H.-J. Hong, M. J. Bibb, and M. J. Buttner. 2002. The ECF 4
sigma factors of Streptomyces coelicolor A3(2). p. 105-125. In D. A. Hodgson 5
and C. M. Thomas (ed.), Signals, Switches, Regulons and Cascades: Control of 6
Bacterial Gene Expression. Cambridge University Press. 7
26. Slavny, P., R. Little, P. Salinas, T. A. Clarke, and R. Dixon. 2010. Quaternary 8
structure changes in a second Per-Arnt-Sim domain mediate intramolecular 9
redox signal relay in the NifL regulatory protein. Mol. Microbiol. 75:61-75. 10
27. Staron, A., H. J. Sofia, S. Dietrich, L. E. Ulrich, H. Liesegang, and T. 11
Mascher. 2009. The third pillar of bacterial signal transduction: classification of 12
the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 13
74:557-81. 14
28. Stein, T., S. Borchert, P. Kiesau, S. Heinzmann, S. Kloss, C. Klein, M. 15
Helfrich, and K. D. Entian. 2002. Dual control of subtilin biosynthesis and 16
immunity in Bacillus subtilis. Mol. Microbiol. 44:403-16. 17
29. Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with 18
apparent streptomycete promoters. Nucleic Acids Res. 20:961-74. 19
30. Tahlan, K., S. K. Ahn, A. Sing, T. D. Bodnaruk, A. R. Willems, A. R. 20
Davidson, and J. R. Nodwell. 2007. Initiation of actinorhodin export in 21
Streptomyces coelicolor. Mol. Microbiol. 63:951-61. 22
23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

25
Figure Legends 1
2
FIG. 1. The microbisporicin gene cluster. mibA encodes the microbisporicin 3
prepropeptide and is co-transcribed with genes encoding biosynthetic enzymes for 4
formation of lanthionine bridges (mibBC) and S-[(Z)-2-aminovinyl]-D-cysteine (mibD), 5
and with genes encoding a putative exporter (mibTU) and a gene of unknown function 6
(mibV). A two-component ABC-transporter encoded by mibEF and a lipoprotein 7
encoded by mibQ may confer immunity to microbisporicin (4). Further modifications are 8
mediated by a tryptophan halogenase-flavin reductase couple (mibHS) and likely by a 9
cytochrome P450 (mibO). Putative regulatory proteins are encoded by mibXW and 10
mibR. mib genes of unknown function are mibJ, mibYZ (possibly encoding an ABC-11
transporter) and mibN (encoding a sodium/proton anti-porter). For a detailed 12
description of mib gene function see (4). The positions of the predicted ECF sigma 13
factor promoter consensus sequences (full arrows) and the expected position of the 14
promoter for mibA (broken arrow) are shown. 15
16
FIG. 2. High-resolution S1 nuclease mapping of the 5’ end of the mibA and mibE 17
transcripts using PCR-generated probes and RNA from wild type M. corallina NRRL 18
30420. The most likely transcription start site is indicated by an asterisk. The full-length 19
probe was run as a control for the identification of full-length protection. The Maxam-20
Gilbert GA chemical sequencing ladder was generated from the full-length probe. The 21
marker was 32P-labelled HinfI digested ΦX174 DNA. For mibE, the length of the 22
protected fragment (determined from the size markers) and the sequencing ladder were 23
used to unambiguously assign the transcriptional start site. 24
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

26
1
FIG. 3. Summary of the results of high-resolution S1 nuclease mapping of the 5’ ends 2
of transcripts from mibJ, mibX, mibR, mibQ and mibE using PCR-generated probes and 3
RNA from wild type M. corallina NRRL 30420. The predicted -35 and -10 elements are 4
conserved and are highlighted with grey boxes. The determined transcriptional start 5
sites (+1) are highlighted in bold. 6
7
FIG. 4. The predicted promoter site of mibA. The transcription start site indicated by S1 8
nuclease mapping is shown (+1) in bold. The predicted -35 and -10 regions are 9
underlined. 10
11
FIG. 5. A bacterial-two hybrid experiment to investigate the interaction between MibW 12
and σMibX. The listed pairs of constructs were transferred into the BACTH reporter strain 13
E. coli BTH101 by transformation. The resulting transformants were selected on 14
MacConkey/Maltose agar containing 100 µg/ml carbenicillin and 25 µg/ml 15
chloramphenicol and incubated at 30°C for several days before two independent clones 16
were picked and subjected to β-galactosidase assays. The histogram was plotted using 17
the average activity of the two clones, in Miller units. Error bars represent the spread of 18
values between clones tested. 19
20
FIG. 6. Luciferase-reporter analysis of σMibX activity in S. coelicolor M1146. pIJ5972 21
(bottom panel) contains promoterless luxAB. In pIJ12341, the mibA promoter (PmibA) 22
transcribes luxAB in the presence of σmibX. pIJ12342 differs by the absence of σmibX. In 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

27
pIJ12343, the mibX promoter (PmibX) transcribes luxAB in the presence of σmibX. 1
pIJ12344 differs by the absence of σmibX. Three independent clones of S. coelicolor 2
M1146 (shown side by side) containing these constructs were grown in duplicate 3
(indicated by numbers 1 and 2 to the left of the images) for 2 d on Difco nutrient agar 4
and light production was visualised using a NightOwl Camera (Berthold) after applying 5
the substrate N-decanal on filter paper discs for 5 minutes. The images shown are 6
representative examples from three independent experiments. 7
8
FIG. 7. Analysis of the effect of replacing mibR with the apr cassette from pIJ773. 9
Mutant (∆R) and wild type (WT) strains of M. corallina NRRL 30420 were grown for 7 d 10
in VSPA medium and 40 µl of culture supernatant assayed for activity against M. luteus. 11
12
FIG. 8. Analysis of the effect of deleting mibX or mibR on mib gene expression. The 13
expression of each gene displayed on the vertical axis was assessed by qRT-PCR in 14
three biological replicates of M. corallina NRRL 30420 ∆mibX::apr (grey histograms) 15
and ∆mibR::apr (black histograms). The number of copies of each transcript was 16
normalised to the number of copies of hrdB in each sample. The normalised copy 17
numbers were averaged for the three biological replicates of each sample and are 18
displayed as a percentage of wild type expression. 19
20
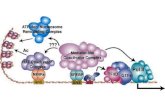
FIG. 9. Model for the regulation of microbisporicin biosynthesis. Prior to detectable 21
microbisporicin production, MibW sequesters σMibX at the membrane, preventing its 22
interaction with target promoters. An unknown signal (indicated by a question mark) 23
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

28
activates transcription of mibR at an unidentified promoter (P?). MibR then activates 1
transcription of the mibABCDTUV operon leading to production of microbisporicin. 2
Interaction of the peptide with MibW, or a low level of inhibition of peptidoglycan 3
biosynthesis that may be perceived by the anti-sigma factor, results in inactivation of 4
MibW, release of σMibX, and high level expression of the entire mib gene cluster. 5
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 1. The microbisporicin gene cluster. mibA encodes the microbisporicin prepropeptide
and is co-transcribed with genes encoding biosynthetic enzymes for formation of
lanthionine bridges (mibBC) and S-[(Z)-2-aminovinyl]-D-cysteine (mibD), and with genes
encoding a putative exporter (mibTU) and a gene of unknown function (mibV). A two-
component ABC-transporter encoded by mibEF and a lipoprotein encoded by mibQ may to
confer immunity to microbisporicin (4). Further modifications are mediated by a tryptophan
halogenase-flavin reductase couple (mibHS) and likely by a cytochrome P450 (mibO).
Putative regulatory proteins are encoded by mibXW and mibR. mib genes of unknown
function are mibJ, mibYZ (possibly encoding an ABC-transporter) and mibN (encoding a
sodium/proton anti-porter). For a detailed description of mib gene function see (4). The
positions of the predicted ECF sigma factor promoter consensus sequences (full arrows)
and the expected position of the promoter for mibA (broken arrow) are shown.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 2. High-resolution S1 nuclease mapping of the 5’ end of the mibA and mibE
transcripts using PCR-generated probes and RNA from wild type M. corallina NRRL
30420. The most likely transcription start site is indicated by an asterisk. The full-length
probe was run as a control for the identification of full-length protection. The Maxam-
Gilbert GA chemical sequencing ladder was generated from the full-length probe. The
marker was 32P-labelled HinfI digested ΦX174 DNA. For mibE, the length of the protected
fragment (determined from the size markers) and the sequencing ladder were used to
unambiguously assign the transcriptional start site.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 3. Summary of the results of high-resolution S1 nuclease mapping of the 5’ ends of
transcripts from mibJ, mibX, mibR, mibQ and mibE using PCR-generated probes and RNA
from wild type M. corallina NRRL 30420. The predicted -35 and -10 elements are
conserved and are highlighted with grey boxes. The determined transcriptional start sites
(+1) are highlighted in bold.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 4. The predicted promoter site of mibA. The transcription start site indicated by S1
nuclease mapping is shown (+1) in bold. The predicted -35 and -10 regions are
underlined.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 5. A bacterial-two hybrid experiment to investigate the interaction between MibW and
σMibX. The listed pairs of constructs were transferred into the BACTH reporter strain E. coli
BTH101 by transformation. The resulting transformants were selected on
MacConkey/Maltose agar containing 100 µg/ml carbenicillin and 25 µg/ml chloramphenicol
and incubated at 30°C for several days before two independent clones were picked and
subjected to β-galactosidase assays. The histogram was plotted using the average activity
of the two clones, in Miller units. Error bars represent the spread of values between clones
tested.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 6. Luciferase-reporter analysis of σMibX activity in S. coelicolor M1146. pIJ5972
(bottom panel) contains promoterless luxAB. In pIJ12341, the mibA promoter (PmibA)
transcribes luxAB in the presence of σmibX. pIJ12342 differs by the absence of σmibX
. In
pIJ12343, the mibX promoter (PmibX) transcribes luxAB in the presence of σmibX. pIJ12344
differs by the absence of σmibX. Three independent clones of S. coelicolor M1146 (shown
side by side) containing these constructs were grown in duplicate (indicated by numbers 1
and 2 to the left of the images) for 2 d on Difco nutrient agar and light production was
visualised using a NightOwl Camera (Berthold) after applying the substrate N-decanal on
filter paper discs for 5 minutes. The images shown are representative examples from three
independent experiments.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 7. Analysis of the effect of replacing mibR with the apr cassette from pIJ773. Mutant
(∆R) and wild type (WT) strains of M. corallina NRRL 30420 were grown for 7 d in VSPA
medium and 40 µl of culture supernatant assayed for activity against M. luteus.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 8. Analysis of the effect of deleting mibX or mibR on mib gene expression. The
expression of each gene displayed on the vertical axis was assessed by qRT-PCR in three
biological replicates of M. corallina NRRL 30420 ∆mibX::apr (grey histograms) and
∆mibR::apr (black histograms). The number of copies of each transcript was normalised to
the number of copies of hrdB in each sample. The normalised copy numbers were
averaged for the three biological replicates of each sample and are displayed as a
percentage of wild type expression.
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from

FIG. 9. Model for the regulation of microbisporicin biosynthesis. Prior to detectable 1
microbisporicin production, MibW sequesters σMibX at the membrane, preventing its 2
interaction with target promoters. An unknown signal (indicated by a question mark) 3
activates transcription of mibR at an unidentified promoter (P?). MibR then activates 4
transcription of the mibABCDTUV operon leading to production of microbisporicin. 5
Interaction of the peptide with MibW, or a low level of inhibition of peptidoglycan 6
biosynthesis that may be perceived by the anti-sigma factor, results in inactivation of 7
MibW, release of σMibX, and high level expression of the entire mib gene cluster. 8
9
on January 5, 2021 by guesthttp://jb.asm
.org/D
ownloaded from