BIOL4800 4801 syllabus...BIOL%4800/4801%Syllabus%Fa.%2014% % % % % % Page%1%of%4%...
Transcript of BIOL4800 4801 syllabus...BIOL%4800/4801%Syllabus%Fa.%2014% % % % % % Page%1%of%4%...

BIOL 4800/4801 Syllabus Fa. 2014
Page 1 of 4
Tools in Microbial Computational Biology BIOL 4800/4801, Fall 2014 When: 1230-‐1320 W, 1230-‐1520 F Where: TBD Instructor: J. Cameron Thrash, Ph.D. Email: [email protected] Twitter handle: @DrJCThrash My office: A112 Life Sciences Annex Prerequisite: General Microbiology BIOL2051 Required readings/websites: The course will require the following books:
Practical Computing for Biologists, Haddock & Dunn Phylogenomics, Desalle & Rosenfeld Course website(s): Moodle eCommunication Policy: The best way to contact me is through email and/or twitter. I will respond to email or twitter messages within 6 hours, except on weekends or between 10pm and 7am. I may respond much quicker, because like you I am glued to my devices, but I do have a life outside of teaching and research (when I’m lucky). If you want an individual physical/video call appointment, email me with a short description of your issue and the desired time and duration of the meeting. This will be subject to my availability. I accept and encourage twitter follows, but I do not accept any other social media friend requests (until graduation). Course description. In modern biology, the need for competence in computational tools is becoming as ubiquitous as that for traditional techniques like PCR. This course will provide basic training in navigating the command-‐line environment, utilizing common tools for genomics and ecology, submitting jobs to High Performance Computing clusters, and managing input and output files. It is NOT a programming class. Prior computational experience is helpful, but not required, as the goal of this course is to bring neophytes to a basic level of competence with common computational biology methods. While the focus will be applying these to microbiological research, many tools are system/organism independent. Classes will take place in a computer lab (TBD) and will have access to the LSU High Performance Computing (HPC) infrastructure. Each week will consist of two hours of theory/practical lecture along with two hours of computer laboratory exercises (plus take-‐home exercises). The 4800/4801 course can be taken for credit by upper-‐level undergraduates and graduate students equally (3 credit hours). Course learning outcomes. By the end of this course, you should be able to:
• Understand a HPC infrastructure • Remotely access a HPC cluster using the command line • Navigate and manipulate the file structure within a Linux environment • Complete basic file manipulation tasks using Linux commands • Write basic shell scripts for parsing input and output files and sending jobs to the compute nodes • Download genomic information from public databases directly to a HPC cluster • Execute parallel (threaded) analyses using BLAST, HMMER, and multiple alignment tools • Understand the modern sequencing platform methodologies, capabilities and limitations • Execute threaded RAxML and MrBayes phylogenetic inferences from multiple alignments • Curate genome sequencing data prior to manipulation • Perform basic automated microbial genome assembly and annotation with the A5 pipeline • Construct orthologous groups from a group of closely related genome-‐sequenced microorganisms • Assess the core and pan-‐genome of a group of closely related microorganisms
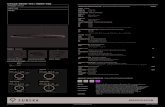
Circular representation of multiple bacterial
genomes (Grote et al. 2012 mBio)

BIOL 4800/4801 Syllabus Fa. 2014
Page 2 of 4
How we’re going to get there (Course Philosophy and Format) Philosophy. This course is designed to get you to a basic working knowledge of many of the common tools used in modern bioinformatics, particularly as applied to microbial genomics. This is a combined lecture/laboratory course, with the laboratory portion spent utilizing computers instead of a typical wet lab. While there will be some lecture component during the Wednesday class, as much as possible this period will have active learning exercises instead of me simply standing around talking. Extensive research on education and the neuroscience of learning has shown there are much more effective ways for us to learn than by sitting and listening to a person stand in the front of the room and talk. You don’t have to come to class to learn that way anyway, for there are endless lectures and resources available online, many from the most eminent scientists in their fields. Some of these will be part of your pre-‐class assignments. Therefore, I endeavor to make class time as effective as possible for stimulating your investment in the material and activating all modes of thinking. The added benefit of being able to do this work yourselves in the lab portion of the class will help complete the process. Classroom mechanics. The one-‐hour Wednesday class will be chiefly concerned with theory and applications behind the various tools we will be learning to use. The three-‐hour Friday class will have a practical lecture for instructional purposes and then two hours of lab time to proceed through exercises designed to help you learn the tools themselves. There will be some assigned readings/podcasts/web-‐videos/lecture slides you will be responsible for at each class period, listed as “Readings, etc.” in the class schedule, below. I will also introduce these pre-‐lecture assignments each week by email. There will be a short online Moodle quiz on the material for Wednesdays that closes one hour before class. Computational Requirements. Our classes will be conducted in a computer lab, TBD. Prior to the first class, you need to have requested an account with LSU HPC for access to the super computer SuperMike II. You will use this account for completing classroom exercises and major assignments. For in-‐class exercises, you will be using the lab computers and logging on through a terminal. For your major assignments, you will need another computer with terminal login capabilities so that you may access SuperMike II remotely. All exercises involving significant computational effort will require the use of a class computing allocation. Details for operating in the HPC environment will be presented during the first two weeks of the course. Major Assignments. In addition to your outputs from classroom exercises you will be responsible for a series of major assignments will be either continuations of the exercises in the Friday lab period, more difficult/comprehensive versions of these exercises, or small team projects that will make use of one or more of the tools we have learned. All major assignments will have a written component that accompanies the results of your computer work and be graded according to a specific rubric. These may include additional reading. You will be graded on the following: Quizzes 10% In-‐class exercises 30% Major Assignments 60%
There will be 1000 total points, graded accordingly: A 900-‐1000 B 800-‐899 C 700-‐799 D 600-‐699 F < 600
Late assignments. Assignments will sacrifice 10% of their points per day they are late. Other important information Absences/Code of Student Conduct. You are expected to have read, understand, and adhere to the LSU Absence Policy (http://saa.lsu.edu/important-‐lsu-‐policies) and the Code of Student Conduct (http://saa.lsu.edu/code-‐student-‐conduct). Our goal should be to learn, not simply to get grades. In science, as in life, your integrity is one of, if not the, most valuable asset you have. Preserve it, protect it, cultivate it.

BIOL 4800/4801 Syllabus Fa. 2014
Page 3 of 4
Students with Disabilities. If anyone has a disability that may require accommodation, you should immediately contact the office of Services for Students with Disabilities to officially document the needed accommodation. The instructor must be presented with this documentation during the first week of class. Time requirements. It is expected that you will have read or viewed the assigned material prior to class for the background necessary to properly participate in the activities and think critically about the concepts addressed. As a general policy, for each hour you are in class, you (the student) should plan to spend at least two hours preparing for the next class. Since this course is for three credit hours, you should expect to spend around six hours outside of class each week reading or working on assignments for the class. Class schedule The schedule is preliminary and subject to change depending on how quickly we are moving through the material. Details on your pre-‐class readings, etc., are supplied below. Class Date Subject Readings, etc. Major Assignment 1 August 27th (W) The command line environment 1, 2, 3, 4, 5, 6 2 August 29th (F) HPC tutorial Complete lab 3 September 3rd (W) Basic Linux commands 7, 8, 9, 10, 11 4 September 5th (F) Basic Linux commands, shell text editors Complete lab 5 September 10th (W) Database access 12 6 September 12th (F) Collecting and manipulating fasta/Genbank files MA 1 7 September 17th (W) Local alignment and dynamic programming 13 8 September 19th (F) BLAST MA 2 9 September 24th (W) Multiple sequence alignment 14 10 September 26th (F) clustalW, MUSCLE, T-‐Coffee MA 3 11 October 1st (W) Genome sequencing 15, 16 October 3rd (F) Fall Break 12 October 8th (W) Curating sequencing output 17 13 October 10th (F) Quality scores, trimming, dereplicating MA 4 14 October 15th (W) Assembly and annotation methods 18, 19 15 October 17th (F) A5 pipeline MA 5 16 October 22nd (W) Hidden Markov Models 20, 21 17 October 24th (F) HMMER MA 6 18 October 29th (W) Applying HMMs-‐ pFam, SFams 22, 23 19 October 31st (F) SFam database searches MA 7 20 November 5th (W) Single gene phylogeny 24, 25 21 November 7th (F) RAxML, MrBayes MA 8 22 November 12th (W) Orthology determination 26 23 November 14th (F) All vs. all BLASTP + OrthoMCL MA 9 24 November 19th (W) Assessing core and pan genomes 27, 28, 29 25 November 21st (F) ITEP MA 10 November 26th (W) Thanksgiving November 28th (F) Thanksgiving 26 December 3rd (W) Presenting MA10 27 December 5th (F) Presenting MA10 Readings, etc. to be completed before class 1. Read the Syllabus 2. Apply for an HPC account (https://accounts.hpc.lsu.edu/login_request.php) 3. Software Carpentry Unix shell tutorials (http://software-‐carpentry.org/v4/shell/index.html)-‐
Introduction, Files and Directories, Creating and Deleting 4. Review the HPC@LSU website (http://www.hpc.lsu.edu), familiarizing yourself with Accounts and
Allocations, the LSU HPC Usage Policy, the User Guide for SuperMike II, and the Computational Biology tools available.
5. Haddock and Dunn, Chp. 4

BIOL 4800/4801 Syllabus Fa. 2014
Page 4 of 4
6. Haddock and Dunn, Chp. 20 7. Haddock and Dunn, Chp. 2 8. Haddock and Dunn, Chp. 3 9. Haddock and Dunn, Chp. 5 10. Software Carpentry Unix shell tutorials (http://software-‐carpentry.org/v4/shell/index.html)-‐ Pipes and
Filters through remaining tutorials. 11. Software Carpentry regex tutorials (http://software-‐carpentry.org/v4/regexp/index.html)-‐ all. 12. DeSalle and Rosenfeld, Chp. 4 13. DeSalle and Rosenfeld, Chp. 5 14. DeSalle and Rosenfeld, Chp. 6 15. Mardis, E. R. (2008). Next-‐Generation DNA Sequencing Methods. Annual Review of Genomics and Human
Genetics, 9(1), 387–402. 16. Metzker, M. L. (2010). Sequencing technologies -‐ the next generation. Nature Reviews Genetics, 11(1),
31–46. 17. TBD 18. Tritt, A., Eisen, J. A., Facciotti, M. T., & Darling, A. E. (2012). An Integrated Pipeline for de Novo Assembly
of Microbial Genomes. Plos One, 7(9), e42304. 19. TBD 20. Eddy, S. R. (2004). What is a hidden Markov model? Nature Biotechnology, 22(10), 1315–1316. 21. Eddy, S. R. (2011). Accelerated Profile HMM Searches. PLOS Computational Biology, 7(10), e1002195. 22. TBD 23. Sharpton, T. J., Jospin, G., Wu, D., Langille, M. G., Pollard, K. S., & Eisen, J. A. (2012). Sifting through
genomes with iterative-‐sequence clustering produces a large, phylogenetically diverse protein-‐family resource. BMC Bioinformatics, 13, 264.
24. DeSalle and Rosenfeld, Chp. 8 25. TBD 26. TBD 27. Benedict, M. N., Henriksen, J. R., Metcalf, W. W., Whitaker, R. J., & Price, N. D. (2014). ITEP: An integrated
toolkit for exploration of microbial pan-‐genomes. BMC Genomics, 15(1), 8. 28. Grote, J., Thrash, J. C., Huggett, M. J., Landry, Z. C., Carini, P., Giovannoni, S. J., & Rappé, M. S. (2012).
Streamlining and Core Genome Conservation among Highly Divergent Members of the SAR11 Clade. mBio, 3(5), e00252–12.
29. TBD Major Assignments (MA) 1. Download the genome sequences for a set of microorganisms from one of the major databases (e.g.,
GenBank, IMG) using the command line, including protein fastas, nucleotide fastas, scaffold fastas, and genbank files. Practice manipulating fasta files with basic linux commands. (50 pts)
2. Execute a threaded BLAST search of a set of proteins with similar annotations against the nr or IMG v4 databases. Curate the results by best hit. (50 pts)
3. Produce threaded multiple-‐sequence alignments using different programs, visualize the alignments with graphical software, compare by eye. (50 pts)
4. Download the raw sequencing data for a microbial genome of choice in GenBank. Dereplicate if necessary, quality trim, and prepare for assembly. (50 pts)
5. Complete an assembly of a microbial genome using A5. (60 pts) 6. Convert your multiple sequence alignments from MA3 into HMMs, search these against a database like nr
or IMG using threaded HMMER call. (60 pts) 7. Search the ORFs from your A5 assembly against the SFam database and curate at the level of a protein
family. Compare with the A5 annotations. (60 pts) 8. Execute threaded phylogenetic inferences using RAxML and MrBayes for 10 of the genes from your
assembly. (60 pts) 9. All vs. all blasts and OrthoMCL for your assembled genome with others that are closely related. (60 pts) 10. Complete core and pan-‐genome analysis of your genome with others that are closely related using ITEP,
reporting the various additional outputs. Format for final presentation to the class. (100 pts)