Bayesian Networks Practice - SNU · Create and modify network models with a graphical editor...
Transcript of Bayesian Networks Practice - SNU · Create and modify network models with a graphical editor...
2016-03-15 Byoung-Hee Kim
Biointelligence Lab, CSE, Seoul National University
Bayesian Networks PracticePart 1
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Agenda
❖ Motivating Example (15 min) ❖ Bayesian Network & Probabilistic Graphical Model (10min)
❖ Tool for Practice: GeNIe (5 min)
❖ Bayesian Network Practice with GeNIe & Weka ■ Main Practices
■ Designing a knowledge system (model) (20 min) ■ Inference with the given model (30 min)
■ Learning Bayesian Network models from data (30 min)
■ Homework & Exercise (20 min)
❖ Appendix: ■ AI & Uncertainty ■ Bayesian Networks in detail
2
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice Outline
3
❖ Designing a Bayesian Network ■ Tool: GeNIe (https://dslpitt.org/genie/, http://www.bayesfusion.com/)
❖ Inference based with a Bayesian Network model ❖ Learning Bayesian Network Models from data
1
2
3
Designing the structure1 Setting Parameters1
Q1?
A1 unlikely
Doing inferences 2
Learning from data3
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 5
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 6
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
Assuming binary states for all the variables Ex) Season: dry or rainy Ex) Sprinkler: ON or OFF
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 7
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 8
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
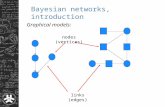
(directed acyclic graph)
: node : edgeparent node child node
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 9
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 10
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 11
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr 12
Slide from K. Mohan & J. Pearl, UAI ’12 Tutorial on Graphical Models for Causal Inference
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Example: Knowledge Engineering
❖ Designing a Bayesian Network: Wet Road ■ My own design of conditional probability tables
13
DRY 0.6
RAINY 0.4
SEASON
SEASON DRY RAINY
ON 0.8 0.1
OFF 0.2 0.9
SEASON DRY RAINY
YES 0.3 0.8
NO 0.7 0.2
WET YES NO
YES 0.8 0.1
NO 0.2 0.9
SPRINKLER RAIN
WET
SLIPPERY
SPRINKLER ON OFF
RAIN YES NO YES NO
YES 0.8 0.1 0.8 0.1
NO 0.2 0.9 0.2 0.9
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
General Concept of Bayesian Networks & Probabilistic Graphical Models
14
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
ACM Turing Award
❖ Nobel Prize in Computing ❖ 2011 Winner: Judea Pearl (UCLA)
■ For fundamental contributions to artificial intelligence through the development of a calculus for probabilistic and causal reasoning
■ Invention of Bayesian networks ■ Pearl's accomplishments have “redefined the term
'thinking machine’” over the past 30 years ■ BN mimics “the neural activities of the human brain,
constantly exchanging messages without benefit of a supervisor”
15
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Bayesian Network as a Probabilistic Graphical Model
16
Probabilistic Graphical model
directed graphundirected graph
C
E
D
BA
C
E
D
BA
Bayesian NetworksMarkov Random Field
Graph
Probability
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Representation of Joint Probability
),,(),,(),(1
)(1),,,,(
321 EDCDCBBA
EDCBAPi
ii
ϕϕϕα
ϕα
=
= ∏ Z
))(|())(|( ))(|())(|())(|(),,,,(
EEPDDPCCPBBPAAPEDCBAP
PaPaPaPaPa=
17
C
E
D
BA
Z3
Z2
Z1
C
E
D
BA
Bayesian Networks
Markov Random Field
The joint probability is effectively represented as the product of independent factors
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Example: Handwriting Recognition
21
[S.-J. Cho, 2003, 2004]
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Applications Examples
❖ APRI system developed at AT&T Bell Labs ■ Fraud/Uncollectible debt detection ■ learns & uses Bayesian networks from data to identify
customers liable to default on bill payments
❖ NASA Vista system ■ predict failures in propulsion systems ■ considers time criticality & suggests highest utility action ■ dynamically decide what information to show
❖ Microsoft Pregnancy and Child-Care ■ Available on MSN in Health section ■ Frequently occurring children’s symptoms are linked to expert
modules that repeatedly ask parents relevant questions ■ Asks next best question based on provided information ■ Presents articles that are deemed relevant based on information provided
22
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Bayesian Network Classifiers
23
Naïve Bayes Classifier Tree-Augmented Naïve Bayes Classifier (TAN)
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Bayesian Network as a Framework
❖ Many probabilistic models can be represented as Bayesian networks ■ Hidden Markov Models ■ Kalman Filters / Particle Filters ■ PCA, ICA, … ■ Hierarchical Bayes models ■ Etc.
24
oTo1 otot-1 ot+1
x1 xt+1 xTxtxt-1
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Designing Bayesian Network Model
❖ TakeHeart II: Decision support system for clinical cardiovascular risk assessment
25
1
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Inference with Bayesian Network Model
❖ Given an assignment of a subset of variables (evidence) in a BN, estimate the posterior distribution over another subset of unobserved variables of interest.
26
Inferences viewed as message passing along the network
2
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Bayesian Network – The Tool for Practice
❖ GeNIe: http://genie.sis.pitt.edu/, http://www.bayesfusion.com/ ■ GeNIe (Graphical Network Interface) is the graphical
interface to SMILE, a fully portable Bayesian inference engine
■ What you can do with GeNIe are ■ Create and modify network models with a graphical editor ■ Building Bayesian networks, influence diagrams ■ Learning structures and parameters of Bayesian networks
from data ■ Probabilistic inference with given Bayesian network
29
Note: SMILE (Structural Modeling, Inference, and Learning Engine) is a fully portable library of C++ classes implementing graphical decision-theoretic methods(Look slide 42 for more information)
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Model developer module: GeNIe. Implemented in Visual C++ in Windows environment. GeNIe
GeNIeRate
SMILE.NET☺Wrappers: SMILE.NET☺ jSMILE☺ , Pocket SMILE☺
Allow SMILE☺ to be accessed from applications other than C++compiler
jSMILE☺Pocket SMILE☺
GeNIe and SMILE☺
30
A developer’s environment for graphical decision models
Reasoning engine: SMILE☺ (Structural Modeling, Inference, and Learning Engine). A platform independent library of C++ classes for graphical models.
SMILE☺
SMiner
Learning and discovery module: SMiner
Support for model building: ImaGeNIe
ImaGeNIe
Diagnosis: Diagnosis
Diagnosis
Qualitative interface: QGeNIe
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice
❖ Designing a Disease-Test Model ■ Building a Bayesian network manually using GeNIe
❖ Inference ■ In GeNIe, try following menus
■ ‘Network – Update Beliefs’, or ‘F5’ ■ Network - Probability of Evidence ■ Network - Annealed MAP
❖ Learning from Data ■ (Data generation using an existing BN model) ■ Learning Bayesian network from data using GeN
■ Structure learning ■ Parameter learning
■ Analyzing the resulting model
31
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Building a Bayesian Network
32
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Building a Bayesian Network
33
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Building a Bayesian Network
34
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Building a Bayesian Network
35
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Building a Bayesian Network
36
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Building a Bayesian Network
37
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 1: Disease-Test Model
38
P(Test=negative | Disease=absent) = 0.95P(Test=negative | Disease=present) = 0.02P(Test=positive | Disease=absent) = 0.05P(Test=positive | Disease=present) = 0.98
P(Disease=absent) = 0.95P(Disease=present) = 0.05
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Disease-Test Model
❖ Inference ■ Set evidence ■ Update belief ■ Check updated
probability
39
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Wet Road Model
40
❖ Open ‘example_wet_road.xdsl’ in GeNIe ❖ Check the structure ❖ Check the parameters
■ Double-click each node and choose the ‘Definition’ tab to see the ‘conditional probability table (CPT)’
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Wet Road Model
41
❖ The first inference ■ Represent each node as a bar chart
■ Check following menu: [Node – View as – Bar Chart] ■ Or just press ‘F8’
■ Inference: get marginalized probabilities for all the nodes ■ Check following menu: [Network – Update Beliefs] ■ Or just press ‘F5’
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Wet Road Model
❖ More basic inferences for Q&A’s ■ Q1: If the season is dry and the
pavement is slippery, did it rain?
❖ Analysis of the question ■ Given evidences: SEASON = DRY,
SLIPPERY = YES ■ Target node to check: RAIN
❖ Inference ■ Set evidences by clicking the given values ■ Press ‘F5’ to update beliefs and check the
updated bar chart of the target and related nodes
❖ Analysis of the inference result ■ A1: Unlikely, it is more likely that the
sprinkler was ON with a very slight possibility that it is not even wet.
42
1
2
1
2
You can see the ‘earth’ sign here
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Wet Road Model
❖ More basic inferences for Q&A’s ■ Q2: But what if we see that the
sprinkler is OFF?
❖ Analysis of the question ■ More evidence: SPRINKLER = OFF ■ Target node to check: RAIN
❖ Inference ■ Set evidences by clicking the given
values ■ Press ‘F5’ to update beliefs and check
the updated bar chart of the target node
❖ Analysis of the inference result ■ A2: Then it is more likely that it
rained
43
1
2
1
2
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Wet Road Model
❖ For each node, we can set following options for various inference ■ Set/clear target ■ Set/clear evidence ■ Control Value
❖ Algorithms for inference in Bayesian networks ■ Exact methods ■ Approximate methods
44
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 2: Inference with the Wet Road Model
❖ Inference: Probability of Evidence, Annealed MAP ■ Based on a set of observed nodes e
■ We can calculate the probability of this observed configuration ■ We can estimate the most probable states of target nodes X
■ Menu: [Network – Probability of Evidence, Annealed MAP] ■ P(e): probability of evidence ■ MAP(maximum a posteriori): maximizing the posterior P(X |e)
45
Q1
A1
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 3: Learning from Data
❖ Learning Bayesian networks using GeNIe
1. In GeNIe, open ‘example_wet_road_5000samples.csv’
2. [Data-Learn New Network] 3. Setting parameters for learning
1. Learning Algorithm: Bayesian Search 2. Max Parent Count: 2 3. Use Accuracy as Score Function. Class Variable: SEASON. 3-
fold cross validation 4. Prior, background knowledge can be
applied by forcing or forbidding edges 5. To apply other learning algorithms, go to
the data matrix by selecting corresponding window in [Window] menu
46
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 3: Learning from Data
❖ Learning Bayesian networks using Weka
1. In Weka Explorer, open ‘example_wet_road_5000samples.csv’
2. In the Classify tab, choose [weka.classifiers.bayes.BayesNet]
3. Set following parameters for BayesNet 1. SearchAlgorithm: K2 2. initAsNaiveBayes: False 3. maxNrOfParents: 2
4. Set ‘Use training set’ as the test option 5. Set ‘SEASON’ as the class label (usually
located at the root node) 6. Click the ‘Start’ button 7. At the generated item in ‘Result list’,
right-click and choose ‘visualize graph’ 8. Ignore the contents in ‘Classifier output’ 9. Click each node to check parameters
47
You can see that the structure and parameters of the original BN were restored by learning
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 3: Learning from Data
❖ Learning Bayesian networks using Weka ■ You may try various search algorithms
■ GeneticSearch ■ HillClimber ■ K2: HillClimber with the fixed order
of variables ■ ..
■ You may try various score metrics ■ BAYES ■ BDeu ■ MDL ■ Entropy ■ AIC
■ You may try various estimators and alpha values
48
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Practice 3: Learning from Data
❖ We can generate samples that follow a Bayesian network model
1. Load a Bayesian network model 2. [Learning – Generate Data File] ■ example_wet_road_5000samples.csv
is a generated file ■ simulation of the wet road model ■ 5,000 instances are sampled from
the wet-road model
49
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Understanding Structure Learning(Score-Based Approaches)
❖ Approach the problem as an optimization problem ❖ Use a scoring metric to measure how well a particular
structure fits the observed set of cases
A B C D
S1 H L L L
S2 H H H H
… … … …
SM L H H L
50
DC
A B
DC
A B
DC
A B
Score
Scoring metric
Search strategy
© 2014-2016, SNU CSE Biointelligence Lab., http://bi.snu.ac.kr
Understanding Parameter Learning
❖ Task: Given a network structure, estimate the parameters of the model from data.
51
SM
S2
S1
LHHL
……..…
HHHH
LLLH
DCBA
A B
C D
P(A)
0.010.99
LH
P(B)
0.070.93
LH
0.60.4(H, H)
0.80.2(H, L)
0.70.3(L, H)
(L, L)
(A, B)
P(C)
0.20.8
LH
0.10.9H
0.90.1L
B
P(D)
LH






























































![Learning Bayesian Networks in R · 2013-07-10 · Bayesian Networks Essentials Bayesian Networks Bayesian networks [21, 27] are de ned by: anetwork structure, adirected acyclic graph](https://static.fdocuments.in/doc/165x107/5f3267ce969e2b02050fd06c/learning-bayesian-networks-in-r-2013-07-10-bayesian-networks-essentials-bayesian.jpg)