Ambrosia trifida) and the Rapid Necrosis Response Horn.pdf(Ambrosia trifida) and the Rapid Necrosis...
Transcript of Ambrosia trifida) and the Rapid Necrosis Response Horn.pdf(Ambrosia trifida) and the Rapid Necrosis...

Glyphosate Resistant Giant Ragweed (Ambrosia trifida) and the Rapid Necrosis Response
PhD student: Christopher Van Horn
Advisor: Dr. Phil Westra
Colorado State University, USA




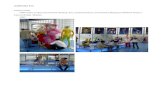
Corn Giant ragweed
Photo: Bill Johnson, University of Iowa

Population Locations
Response to glyphosate:
Red - Susceptible
Green - Resistant Slow Response
Yellow - Resistant Rapid Necrosis

Multiple Sequence Alignment
• Giant ragweed DNA sequences were aligned to a reference EPSPS exon 2 sequence.
• Results show no nucleotide mutations in this binding region of the EPSPS enzyme across all accessions sequenced.

EPSPS Expression Analysis
• No giant ragweed EPSPS signal was detected on the western blot.
• These results suggest that EPSPS is not highly expressed in giant ragweed.
palmer amaranth kochia giant ragweed
R R S R R R R S R R S

Glyphosate Mode of Action • Glyphosate inhibits EPSPS at the sixth step of the shikimic
acid pathway, leading to the accumulation of shikimate.

Shikimate Assay • Leaf discs sampled from untreated plants were
treated with glyphosate.
ng
shik
imat
e /
μl
μM glyphosate
Shikimate Accumulation

Light vs Dark: Resistant Rapid Necrosis
24 hours in light Avg. shikimate accumulation
172 ng /ul
24 hours in dark Avg. shikimate accumulation
28 ng /ul
• Plants treated at 0.9 kg ae ha-1 glyphosate.
24 HAT Dark
24 HAT Light
Hypothesis: Rapid necrosis is a light dependent process.

Light vs Dark: Sucrose • Roots washed and placed in a 2% sucrose solution 10 minutes
prior to glyphosate treatment at 0.9 kg ae ha-1.
• Plants showed the rapid necrosis response in the absence of light when there was an alternative source of carbon available.
Dark Light
24 HAT
Hypothesis: Rapid necrosis is a carbon dependent process.

Light vs Dark: Sucrose • Leaf discs were sampled from 0.9 kg ae ha-1 glyphosate treated
resistant rapid necrosis plants 24 HAT.
• Glyphosate treated plants accumulated shikimate in the dark when sucrose was supplied.
0
50
100
150
200
250
300
350
400
Young Leaf Old Leaf Young Leaf Old Leaf
Light Dark
ng
Shik
imat
e /
ul
Shikimate Accumulation: Light vs Dark with 2% Sucrose
Treated
Untreated
Glyphosate Treated Untreated
ng
shik
imat
e /
ul

Root Absorbed Response to Shikimic Acid • Roots washed and place in 5 mM shikimic acid.
• Rapid necrosis did not occur.
0 HAT 24 HAT
Hypothesis: The accumulation of shikimate does not
directly cause rapid necrosis.

Root Absorbed Response to Salicylic Acid • Roots washed and placed in 5 mM salicylic acid.
• Rapid necrosis did occur in both R and S plants.
0 HAT Resistant
24 HAT Resistant
24 HAT Susceptible
Hypothesis: Glyphosate induces salicylic acid production in
the resistant rapid necrosis biotype.

Root Absorbed Response to Amino Acids • Plant roots washed and placed in solution 10 minutes prior
to 0.9 kg ae ha-1 glyphosate treatment.
• Rapid necrosis did not occur when both phenylalanine and tyrosine were provided.
Phe + Tyr Phenylalanine Tyrosine Water
0 HAT
Phe + Tyr Phenylalanine Tyrosine Water
24 HAT
Hypothesis: Both phenylalanine and tyrosine play a
role in the rapid necrosis response.

Resistant Rapid Necrosis Response
• Only the top leaf was sprayed with 1.8 kg ae ha-1 glyphosate.
10 minutes after treatment 48 hours after treatment
Treated Leaf Treated Leaf

Single Leaf Response
• Excised leaves were placed in 11.8 mM glyphosate.
• Rapid necrosis did not occur.
6 HAT

Shoot and Leaf Response • Excised shoots were placed in 11.8 mM glyphosate.
• Rapid necrosis did occur.
0 HAT
24 HAT
Hypothesis: A signal from meristematic tissue is
required for rapid necrosis.

Conclusions
• Rapid necrosis is dependent on a carbon source.
• Salicylic acid is sufficient to cause rapid necrosis.
• Supplemental phenylalanine and tyrosine prior to glyphosate treatment prevent rapid necrosis.
• Meristematic or shoot tissue is required for rapid necrosis.
• The mechanism of glyphosate resistance is unknown.

Future Research: RNA-seq
• Reference transcriptome: assembled de-novo using RNA sample from a rapid necrosis plant before and after treatment.
• RNA-seq: Sequence 4 resistant populations and 2 susceptible populations. 3 biological replicates from each population.
• Treatment time-points at 15, 30, 60, and 180 MAT.
• Aim: identify genes that are responding to glyphosate and investigate known genes associated with the HR pathway.
Genotype Tissue
collection and RNA extraction
cDNA library construction
and sequencing
Bioinformatics pipeline
Resistant Susceptible

Acknowledgements
• Graduate students Andrew Wiersma – CSU Darci Giacomini – CSU Taylor Jeffery – UG, Ontario Courtney Glettner – UW, Madison The CSU Weed Research Lab
• Collaborators Dale Shaner – USDA-ARS Doug Sammons – Monsanto Steve Weller – Purdue University Burkhard Schulz – Purdue University Chris Hall – University of Guelph Peter Sikkima – University of Guelph Francois Tardif – University of Guelph Kassim AlKhatib – University of California, Davis Dave Stoltenberg – University of Wisconsin, Madison Chris Preston – University of Adelaide, Australia Cecil Stushnoff Frank Dayan – USDA-ARS Fritz Breitenbach – University of Minnesota Stevan Knezevic – University of Nebraska-Lincoln Roland Beffa – Bayer Crop Science Sascha Gille – Bayer Crop Science
• Colorado State University Phil Westra Anireddy Reddy Jan Leach Cris Argueso Scott Nissen Todd Gaines

Questions?