3. DNA, RNA and Protein Synthesis
-
Upload
susan-sylvianingrum -
Category
Documents
-
view
219 -
download
0
Transcript of 3. DNA, RNA and Protein Synthesis
-
7/28/2019 3. DNA, RNA and Protein Synthesis
1/73
LOGO
3. DNA, RNA and Protein
Synthesis
-
7/28/2019 3. DNA, RNA and Protein Synthesis
2/73
Where it all began
You started as a cell smaller thana period at the end of a sentence
.
-
7/28/2019 3. DNA, RNA and Protein Synthesis
3/73
How did you
get from there
to here?
And now look at you
-
7/28/2019 3. DNA, RNA and Protein Synthesis
4/73
Cell cycle & cell division
Cell cycle life of a cell from
origin to division into
2 new daughter cells
Cell division
continuity of life =
reproduction of cells
reproduction unicellular organisms
growth
repair & renew
-
7/28/2019 3. DNA, RNA and Protein Synthesis
5/73
Dividing cell
Dividing cell duplicates DNA separates each copy to
opposite ends of cell
splits into 2 daughter cells
human cell duplicates ~3 meters DNA
separates 2 copies so each daughter cell has
complete identical copy
error rate = ~1 per 100 million bases
-
7/28/2019 3. DNA, RNA and Protein Synthesis
6/73
DNA Replication
The double helix unwinds into 2separate strands. The two
strands serve as templates for
copying two new strands of
DNA each new helix
contains 1 original strand plus 1new strand semi-
conservative replication with
normal base pairing: A with T,
and G with C.
The energy for this comes from
the nucleotide precursors.
They all have 3 phosphates on
them, like ATP, and 2 of the
phosphates are removed tomake the DNA.
-
7/28/2019 3. DNA, RNA and Protein Synthesis
7/73
Nucleotide precursors
ATPAdenosine triphosphate
++
modified nucleotide
adenine ribose + Pi + Pi + Pi
-
7/28/2019 3. DNA, RNA and Protein Synthesis
8/73
Bacterial chromosome replication
The separation ofcomplementary strands
occurs in the regions of
the DNA called origins
of replication (ori).
Bacterial chromosome
have a single origin.
The separated strandsform replication fork
-
7/28/2019 3. DNA, RNA and Protein Synthesis
9/73
Eukaryotic chromosome replication
Because of their large size, eukaryoticchromosome have multiple origins
Replication begins at specific sites
where the two parental strands
separate and form replication
bubbles.
The bubbles expand laterally, as
DNA replication proceeds in both
directions.
Eventually, the replication
bubbles fuse, and synthesis of
the daughter strands is
complete.
1
2
3
Origin of replication
Bubble
Parental (template) strand
Daughter (new) strand
Replication fork
Two daughter DNA molecules
In eukaryotes, DNA replication begins at many sites along the giant
DNA molecule of each chromosome.In this micrograph, three replication
bubbles are visible along the DNA of
a cultured Chinese hamster cell (TEM).
(b)(a)
0.25 m
-
7/28/2019 3. DNA, RNA and Protein Synthesis
10/73
DNA Replication
DNA gyrase
Addition of RNA primer (10-15 bp),
are synthesized by primase
-
7/28/2019 3. DNA, RNA and Protein Synthesis
11/73
Antiparallel Elongation
DNA polymerases cannot initiate the synthesis of apolynucleotide
They can only add nucleotides to the 3 end (5 3)
The initial nucleotide strand
is a primer = RNA sequences, are synthesized byprimase
3
5 3
RNA primer
newly synthesized DNA
5
5
-
7/28/2019 3. DNA, RNA and Protein Synthesis
12/73
RNA primer
5
3
3
5
3
5
direction of leading strand synthesis
direction of lagging strand synthesis
replication fork
Synthesis of the strands
-
7/28/2019 3. DNA, RNA and Protein Synthesis
13/73
5
3 5
3
Movement of the replication fork
Synthesis of the strands
-
7/28/2019 3. DNA, RNA and Protein Synthesis
14/73
Movement of the replication fork
RNA primer
Okazaki fragment
RNA primer
5
Synthesis of the strands
-
7/28/2019 3. DNA, RNA and Protein Synthesis
15/73
3
RNA primer
5
DNA polymerase III initiates at the primer andelongates DNA up to the next RNA primer
5
53
5
newly synthesized DNA (100-1000 bases)
(Okazaki fragment)
53
DNA polymerase I inititates at the end of the Okazaki fragment
and further elongates the DNA chain while simultaneously
removing the RNA primer with its 5 to 3 exonuclease activity
pol III
pol I
-
7/28/2019 3. DNA, RNA and Protein Synthesis
16/73
newly synthesized DNA
(Okazaki fragment)5
3
53
DNA ligase seals the gap by catalyzing the formation
of a 3, 5-phosphodiester bond in an ATP-dependent reaction
Synthesis of the strands
St d ti t th li ti f k iti
-
7/28/2019 3. DNA, RNA and Protein Synthesis
17/73
5
3
3
5
3
5
Strand separation at the replication fork causes positive
supercoiling of the downstream double helix
DNA gyrase is a topoisomerase II, which
breaks and reseals the DNA to introduce negative
supercoils ahead of the fork
Fluoroquinolone antibiotics target DNA gyrases in many
gram-negative bacteria: ciprofloxacin and levofloxacin (Levaquin)
-
7/28/2019 3. DNA, RNA and Protein Synthesis
18/73
Figure 16.16
Overall direction of replicationLeading
strand
Lagging
strand
Lagging
strand
Leading
strandOVERVIEW
Leading
strand
Replication fork
DNA pol III
Primase
PrimerDNA pol III Lagging
strand
DNA pol I
Parental DNA
5
3
43
2
Origin of replication
DNA ligase
1
5
3
Helicase unwinds theparental double helix.1
Molecules of single-
strand binding protein
stabilize the unwound
template strands.
2 The leading strand is
synthesized continuously in the
5 3 direction by DNA pol III.
3
Primase begins synthesis
of RNA primer for fifth
Okazaki fragment.
4
DNA pol III is completing synthesis of
the fourth fragment, when it reaches the
RNA primer on the third fragment, it will
dissociate, move to the replication fork,
and add DNA nucleotides to the 3 end
of the fifth fragment primer.
5 DNA pol I removes the primer from the 5 end
of the second fragment, replacing it with DNA
nucleotides that it adds one by one to the 3 end
of the third fragment. The replacement of the
last RNA nucleotide with DNA leaves the sugar-
phosphate backbone with a free 3 end.
6 DNA ligase bonds
the 3 end of the
second fragment to
the 5 end of the first
fragment.
7
A summary of DNA replication
-
7/28/2019 3. DNA, RNA and Protein Synthesis
19/73
Proteins That Assist DNA Replication
Table 16.1
-
7/28/2019 3. DNA, RNA and Protein Synthesis
20/73
Proteins That Assist DNA Replication :
DNA Polymerase
Prokaryote FunctionPol I Gap filling & DNA repair
Pol II Gap filling & DNA repair
Pol III Replication
Eukaryote
Pol Lagging strand replication
Pol Gap filling & DNA repair
Pol Leading strand replication
Pol Mitochondria replication
Pol Gap filling & DNA repair
-
7/28/2019 3. DNA, RNA and Protein Synthesis
21/73
Eukaryotic chromosomal DNA molecules Have at their ends nucleotide sequences, called
telomeres, that postpone the erosion of genes nearthe ends of DNA molecules
If the chromosomes of germ cells becameshorter in every cell cycle Essential genes would eventually be missing from the
gametes they produce
Figure 16.19 1 m
Replicating the Ends of DNA Molecules
-
7/28/2019 3. DNA, RNA and Protein Synthesis
22/73
Replicating the Ends of DNA Molecules
The ends ofeukaryotic
chromosomal DNA Get shorter
with each
round of
replication
An enzyme calledtelomerase
Catalyzes thelengthening of
telomeres in germcells
Figure 16.18
End of parentalDNA strands Leading strandLagging strand
Last fragment Previous fragment
RNA primer
Lagging strand
Removal of primers and
replacement with DNA
where a 3 end is available
Primer removed but
cannot be replaced
with DNA because
no 3 end available
for DNA polymerase
Second round
of replication
New leading strand
New lagging strand 5
Further rounds
of replication
Shorter and shorter
daughter molecules
5
3
5
3
5
3
5
3
3
-
7/28/2019 3. DNA, RNA and Protein Synthesis
23/73
From nucleus to cytoplasm
Where are the genes? genes are on chromosomes in nucleus
Where are proteins synthesized?
proteins made in cytoplasm by ribosomes How does the information get from nucleus to
cytoplasm?
messenger RNA
nucleus
C l D
-
7/28/2019 3. DNA, RNA and Protein Synthesis
24/73
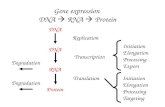
Central Dogma
RNA
DNA
Protein
Transcription
Translation
Replication
Reverse
Transcription
Juang RH (2004) BCbasics
-
7/28/2019 3. DNA, RNA and Protein Synthesis
25/73
RNA
ribose sugar N-bases
uracil instead of thymine,
which U : A modified bases
single stranded
RNADNAtranscription
-
7/28/2019 3. DNA, RNA and Protein Synthesis
26/73
RNA polynucleotide chain
2 -OH makes3, 5 phosphodiester
bond unstable
DNA polynucleotide chain
-
7/28/2019 3. DNA, RNA and Protein Synthesis
27/73
Examples of modified bases found in RNA
Dihydrouridine Pseudouridine 1-methylguanosine 7-methylguanosine
1-methyladenosine 2-thiocytidine 5-methylcytidine Ribothymine
-
7/28/2019 3. DNA, RNA and Protein Synthesis
28/73
Types of RNA
In prokaryote cells :
- messenger RNA (mRNA) :
an exact copy of a gene
- ribosomal RNA (rRNA) :
short (1,500-4,700
nucleotides), components of
ribosome- transfer RNA (tRNA) :
molecule that transport
amino acids to the ribosome
during synthesis protein
In eukaryote cells
-
7/28/2019 3. DNA, RNA and Protein Synthesis
29/73
Transcription
Transcription Is the synthesis of RNA under the direction of DNA
Occurs only in a chromosome segments thatcontain genes, many copies
Transcribed DNA strand = template strand=antisense strand untranscribed DNA strand = coding strand= sense
strand
Synthesis of complementary RNA strand transcription bubble
RNA polymerase : unwinds the DNA helix and thencopies one strand of DNA to RNA
Template ( ) Strand Transcribes to mRNA
-
7/28/2019 3. DNA, RNA and Protein Synthesis
30/73
Template (-) Strand Transcribes to mRNA
3 5
5 3 RNA
template strand
nontemplate strand (+) strand
(-) strand
5 3
coding strand
AUG
Codon is denoted on mRNA
ATG
TAC
UAG
TAG
ATC
Startcodon
Identicalsequence
Juang RH (2004) BCbasics
-
7/28/2019 3. DNA, RNA and Protein Synthesis
31/73
Transcription
Transcription bubble
-
7/28/2019 3. DNA, RNA and Protein Synthesis
32/73
MEKANISME TRANSKRIPSI
= aktivator, o=operator, p=promotor,tsp=situs inisiasi transkripsi, RBS=ribosom
binding site, ATG=kodon start, STOP=kodonstop, t=terminator
Komponen transkripsi : promotor, situs
inisiasi transkripsi, dan terminator
-
7/28/2019 3. DNA, RNA and Protein Synthesis
33/73
MEKANISME TRANSKRIPSI
Promotor : tempat RNA pol mengikatkan diri utkmemulai transkripsi (recognition site) menentukan kecepatan transkripsi
Terdiri dari 2 blok urutan nukleotida yg terpisah :
- blok 1 : ~6 pb, - 5 sampai - 8 (Pribnow box/TATA
box) dari situs inisiasi transkripsi- blok 2 : ~10 pb, - 35 dari situs inisiasi transkripsi
Situs inisiasi transkripsi : tempat awal transkripsi(+1).
Pengenalan & pengikatan RNA pol di blok 2membuat blok 1 membuka mjd untai tunggal.
Salah satu untai tunggal menjadi templat dantranskripsi dimulai di +1.
-
7/28/2019 3. DNA, RNA and Protein Synthesis
34/73
-
7/28/2019 3. DNA, RNA and Protein Synthesis
35/73
Transcription in Prokaryotes
Promoter sequences
RNA polymerase
molecules bound to
bacterial DNA
-
7/28/2019 3. DNA, RNA and Protein Synthesis
36/73
Transcription in Prokaryotes
Initiation RNA polymerase binds to promoter sequence
on DNA
-
7/28/2019 3. DNA, RNA and Protein Synthesis
37/73
Transcription in Prokaryotes
Elongation RNA polymerase unwinds DNA
~20 base pairs at a time
reads DNA 3 5
builds RNA 5 3 (the energy governs the synthesis!)
No proofreading
1 error/105 bases many copies
short life
-
7/28/2019 3. DNA, RNA and Protein Synthesis
38/73
Transcription in Prokaryotes
Termination RNA polymerase stops at termination
sequence
mRNA leaves nucleus through pores
RNA GC
hairpin turn
-
7/28/2019 3. DNA, RNA and Protein Synthesis
39/73
Structure of prokaryotic mRNA
5
3
PuPuPuPuPuPuPuPu AUGShine-Dalgarno sequence initiation
The Shine-Dalgarno (SD) sequence base-pairs with a pyrimidine-rich
sequence in 16S rRNA to facilitate the initiation of protein synthesis
AAUtermination
translated region
T i i i E k
-
7/28/2019 3. DNA, RNA and Protein Synthesis
40/73
LOGO
Transcription in Eukaryotes
-
7/28/2019 3. DNA, RNA and Protein Synthesis
41/73
Prokaryote vs Eukaryote genes
Prokaryotes DNA in cytoplasm
circular chromosome
naked DNA
no introns
Eukaryotes DNA in nucleus
linear chromosomes
DNA wound on histone
proteins introns vs. exons
eukaryotic
DNA
exon = coding (expressed) sequence
intron = noncoding (in between) sequence
Intron and Exon in Eukaryotic Cells
-
7/28/2019 3. DNA, RNA and Protein Synthesis
42/73
Intron and Exon in Eukaryotic Cells
mRNA
DNA
5 3
cap
poly Atail
exon exonexonintron intron
mature mRNA
Processing
Transcription
Splicing
promotor3 5
Take place in nucleus
start codon stop codon
To cytoplasm
Intron deleted
Juang RH (2004) BCbasics
-
7/28/2019 3. DNA, RNA and Protein Synthesis
43/73
Transcription in Eukaryotes
Promoters signal theinitiation of RNA synthesis
Transcription factors Help eukaryotic RNA
polymerase recognize promoter
sequences Initiation complex
transcription factors bind topromoter region upstream ofgene
proteins which bind to DNA &turn on or off transcription
TATA box binding site
only then does RNApolymerase bind to DNA
-
7/28/2019 3. DNA, RNA and Protein Synthesis
44/73
Transcription initiation
Control regions on DNA promoter
nearby control sequence on DNA
binding of RNA polymerase & transcription factors
base rate of transcription
enhancers distant control
sequences on DNA
binding of activatorproteins
enhanced rate (high level)of transcription
-
7/28/2019 3. DNA, RNA and Protein Synthesis
45/73
Transcription in Eukaryotes
3 RNA polymerase enzymes RNA polymerase I
only transcribes rRNA genes
RNA polymerase II transcribes genes into mRNA
RNA polymerase III
only transcribes rRNA genes
each has a specific promoter sequence it
recognizes
P i i l i
-
7/28/2019 3. DNA, RNA and Protein Synthesis
46/73
Post-transcriptional processing
Eukaryotic cells modify RNA after transcription
Enzymes in the eukaryotic nucleus Modify pre-mRNA in specific ways before the genetic
messages are dispatched to the cytoplasm
Modify pre-mRNA (primary transcript) :
- Protect mRNAfrom RNase enzymes in cytoplasm by alteration of mRNA ends
add 5' cap
add polyA tail
A modified guanine nucleotideadded to the 5 end
50 to 250 adenine nucleotidesadded to the 3 end
Protein-coding segment Polyadenylation signal
Poly-A tail3 UTRStop codonStart codon
5 Cap 5 UTR
AAUAAA AAAAAA
TRANSCRIPTION
RNA PROCESSING
DNA
Pre-mRNA
mRNA
TRANSLATIONRibosome
Polypeptide
G P P P
5 3
-
7/28/2019 3. DNA, RNA and Protein Synthesis
47/73
Add 5 Cap
5 cap plays a role in ribosomerecognition of the 5 end of the RNA
molecule during translation
P l d l ti
-
7/28/2019 3. DNA, RNA and Protein Synthesis
48/73
Polyadenylation
Polyadenylation : a string of adenine nucleotides around100- 300 nucleotides in length is added to the 3 end of
the mRNA
This tail protects mRNA from degradation in cytoplasm,
increasing its stability and availability for translation
A modified guanine nucleotideadded to the 5 end
50 to 250 adenine nucleotidesadded to the 3 end
Protein-coding segment Polyadenylation signal
Poly-A tail3 UTRStop codonStart codon
5 Cap 5 UTR
AAUAAA AAAAAA
TRANSCRIPTION
RNA PROCESSING
DNA
Pre-mRNA
mRNA
TRANSLATIONRibosome
Polypeptide
G P P P
5 3
P t t i ti l i
-
7/28/2019 3. DNA, RNA and Protein Synthesis
49/73
Post-transcriptional processing
RNA splicing Removes introns and joins exons
Figure 17.10
TRANSCRIPTION
RNA PROCESSING
DNA
Pre-mRNA
mRNA
TRANSLATION
Ribosome
Polypeptide
5 Cap
Exon Intron
1
5
30 31
Exon Intron
104 105 146
Exon 3Poly-A tail
Poly-A tail
Introns cut out and
exons spliced togetherCoding
segment
5 Cap
1 1463 UTR3 UTR
Pre-mRNA
mRNA
RNA li i
-
7/28/2019 3. DNA, RNA and Protein Synthesis
50/73
RNA splicing
Is carried out byspliceosomes in some cases
antibodies to snRNPs are
seen in the autoimmune
disease systemic lupus
erythematosus (SLE)
Ribozymes
Are catalytic RNA
molecules that function
as enzymes and can
splice RNA
Figure 17.11
RNA transcript (pre-mRNA)
Exon 1 Intron Exon 2
Other proteins
Protein
snRNA
snRNPs
Spliceosome
Spliceosome
components
Cut-out
intronmRNA
Exon 1 Exon 2
5
5
5
1
2
3
The Functional and Evolutionary Importance of
-
7/28/2019 3. DNA, RNA and Protein Synthesis
51/73
y p
Introns
The presence of introns Allows for alternative RNA splicing
Variable processing of exons creates a family of proteins, exp.Antibodies, neurotrasmitter
F t t i
-
7/28/2019 3. DNA, RNA and Protein Synthesis
52/73
mRNA
From gene to protein
DNAtranscription
nucleuscytoplasm
mRNA leavesnucleus throughnuclear pores
proteins synthesized
by ribosomes usinginstructions on mRNA
a
a
a
a
a
a
a
a
a
a
a
a
a
a
aa
ribosome
proteintranslation
H d RNA d f t i ?
-
7/28/2019 3. DNA, RNA and Protein Synthesis
53/73
How does mRNA code for proteins?
TACGCACATTTACGTACGCGGDNA
AUGCGUGUAAAUGCAUGCGCCmRNA
MetArgValAsnAlaCysAlaprotein
?
How can you code for 20 amino acids
with only 4 nucleotide bases (A,U,G,C)?
C ki th d
-
7/28/2019 3. DNA, RNA and Protein Synthesis
54/73
Cracking the code
Nirenberg & Matthaei determined 1st codonamino acid match
UUU coded forphenylalanine
created artificial poly(U) mRNA
added mRNA to test tube of
ribosomes, tRNA & amino acids
mRNA synthesized single
amino acid polypeptide chain
1960 | 1968
phephephephephephe
How are the codons matched to amino
-
7/28/2019 3. DNA, RNA and Protein Synthesis
55/73
acids?
TACGCACATTTACGTACGCGGDNA
AUGCGUGUAAAUGCAUGCGCCmRNA
aminoacid
tRNA
anti-codon
codon
5' 3'
3' 5'
3' 5'
UAC
Met GCA
ArgCAU
Val
-
7/28/2019 3. DNA, RNA and Protein Synthesis
56/73
-
7/28/2019 3. DNA, RNA and Protein Synthesis
57/73
tRNA structure
-
7/28/2019 3. DNA, RNA and Protein Synthesis
58/73
tRNA structure
Clover leaf structure
anticodon on clover leaf end and amino acid attached on
3' end
consists of a single RNA strand that is only about 80
nucleotides long
Loading tRNA
-
7/28/2019 3. DNA, RNA and Protein Synthesis
59/73
Loading tRNA
Aminoacyl tRNAsynthetase
enzyme which bondsamino acid to tRNA
endergonic reaction ATP AMP
energy stored intRNA-amino acid bond
unstable
so it can release aminoacid at ribosome
Ribosomes
-
7/28/2019 3. DNA, RNA and Protein Synthesis
60/73
Ribosomes
Function protein production
Structure
ribosomes contain rRNA & protein
composed of 2 subunits that combine to carryout protein synthesis
P k t k t ib
-
7/28/2019 3. DNA, RNA and Protein Synthesis
61/73
Prokaryote vs. eukaryote ribosomes
Ribosomes
-
7/28/2019 3. DNA, RNA and Protein Synthesis
62/73
Ribosomes
Facilitate coupling oftRNA anticodon tomRNA codon
P site (peptidyl-tRNA site) holds tRNA carrying growing
polypeptide chain A site (aminoacyl-tRNA site)
holds tRNA carrying nextamino acid to be added tochain
E site (exit site) empty tRNAleaves ribosome
from exit site
Translation
-
7/28/2019 3. DNA, RNA and Protein Synthesis
63/73
Translation
Translation Is the actual synthesis of a
polypeptide, which occursunder the direction ofmRNA
Occurs on ribosomes
Codons blocks of 3 nucleotides
decoded intothe sequence of aminoacids
Codons must be read inthe correct reading frame For the specified
polypeptide to be produced
MEKANISME TRANSLASI
-
7/28/2019 3. DNA, RNA and Protein Synthesis
64/73
MEKANISME TRANSLASI
= aktivator, o=operator, p=promotor,tsp=situs inisiasi transkripsi, RBS=ribosom
binding site, ATG=kodon start, STOP=kodonstop, t=terminator
Komponen translasi : kodon start, RBS(ribosom binding site) dan kodon stop
TRANSLASI
-
7/28/2019 3. DNA, RNA and Protein Synthesis
65/73
TRANSLASI
Translasi berlangsung dari aa pada ujung N(amino), berakhir di ujung C (karboksil)
Cetakan : mRNA, ditranslasi dari 5 ke 3
tRNA dan rRNA tidak ditranslasi, tetapi
berfungsi langsung
RBS pada mRNA berikatan dengan ribosom (di
sitoplasma)
Building a polypeptide
-
7/28/2019 3. DNA, RNA and Protein Synthesis
66/73
Building a polypeptide
Initiation brings together mRNA,
ribosome subunits,proteins (initiation factors) &initiator tRNA
Elongation
Termination
Elongation: growing a polypeptide
-
7/28/2019 3. DNA, RNA and Protein Synthesis
67/73
Elongation: growing a polypeptide
catalyzed by peptidyl
transferase
peptydil tRNA
-
7/28/2019 3. DNA, RNA and Protein Synthesis
68/73
Translation in Prokaryotes
-
7/28/2019 3. DNA, RNA and Protein Synthesis
69/73
Translation in Prokaryotes
Transcription & translation are simultaneous in
bacteria
DNA is in
cytoplasm
no mRNA
editing needed
Translation in Prokaryotes
-
7/28/2019 3. DNA, RNA and Protein Synthesis
70/73
Translation in Prokaryotes
Polyribosomes : anumber of ribosomes
can translate a single
mRNA molecule
simultaneously
Figure 17.20a, b
Growing
polypeptides
Completed
polypeptide
Incoming
ribosomal
subunits
Start of
mRNA
(5 end)
End of
mRNA
(3 end)
An mRNA molecule is generally translated simultaneously
by several ribosomes in clusters called polyribosomes.
(a)
Ribosomes
mRNA
This micrograph shows a large polyribosome in a prokaryoticcell (TEM).
0.1 m
(b)
Translation in Eukaryotes
-
7/28/2019 3. DNA, RNA and Protein Synthesis
71/73
Translation in Eukaryotes
In a eukaryotic cell The nuclear
envelope separates
transcription from
translation
Extensive RNA
processing occurs in
the nucleus
Protein Folding and Post-TranslationalM difi ti
-
7/28/2019 3. DNA, RNA and Protein Synthesis
72/73
Modifications
After translation : Proteins may be modified in ways that affect
their three-dimensional shape
Proteins destined for the endomembrane
system or for secretion
Must be transported into the ER
Have signal peptides to which a signal-
recognition particle (SRP) binds, enabling the
translation ribosome to bind to the ER
The signal mechanism for targeting proteins to the ER
-
7/28/2019 3. DNA, RNA and Protein Synthesis
73/73
Ribosome
mRNA
Signal
peptide
Signal-
recognition
particle
(SRP) SRPreceptor
protein
Translocation
complex
CYTOSOL
Signal
peptide
removed
ER
membrane
Protein
ERLUMEN
g g g p
Polypeptide
synthesis begins
on a free
ribosome in
the cytosol.
1 An SRP binds
to the signal
peptide, halting
synthesis
momentarily.
2 The SRP binds to a
receptor protein in the ER
membrane. This receptor
is part of a protein complex
(a translocation complex)
that has a membrane pore
and a signal-cleaving enzyme.
3 The SRP leaves, and
the polypeptide resumes
growing, meanwhile
translocating across the
membrane. (The signal
peptide stays attached
to the membrane.)
4 The signal-
cleaving
enzyme
cuts off the
signal peptide.
5 The rest of
the completed
polypeptide leaves
the ribosome and
folds into its final
conformation.
6